Predesigned siRNA
MISSION® Predesigned siRNA were created using the proprietary Rosetta Inpharmatics siRNA Design algorithm in an exclusive partnership with Merck & Co. The Rosetta siRNA Design Algorithm utilizes Position-Specific Scoring Matrices and knowledge of the seed region to predict the most specific and effective sequences for your target genes. The algorithm’s rules were developed utilizing empirical data collected from gene silencing experiments carried out over three years.
Product Benefits
- Best-in-class, guaranteed gene silencing
- Efficient knockdown of low abundance messages
- Simplified transfection optimization with 11 Positive Control siRNA
- Distinguish sequence-specific silencing from non–specific effects with 8 negative control siRNA
- Hundreds of functionally-validated predesigned siRNA
Product Features
- Species: Human, Mouse & Rat
- Quantities: 2 (10 nmol), 5 (25 nmol) & 10 (50 nmol) OD
- Purification: Desalt or HPLC
- Sequence Form: 21mer duplexes with dTdT overhangs
- Quality Control: 100% mass spectrometry*
- Format: Supplied dry in tubes
*Depending on manufacturing site, PAGE may be used to assess siRNA duplexes.
Product Pools
A popular pooled format is 4 duplexes at 5 nmol each combined into one tube (20 nmol pooled) plus the exact same 4 duplexes also at 5 nmol each in separate tubes (another 20 nmol individual). However, our sophisticated liquid handlers allow for a wide range of other possibilities. For feasibility review of your specific needs, contact sirnarequest@milliporesigma.
Validated siRNA
Many common gene targets have been validated for ≥75% mRNA knockdown (Figure 1 for example data and the table for a list of commonly-ordered, validated siRNA by gene symbol). Validated siRNA are suitable for transfection optimization and as positive controls.
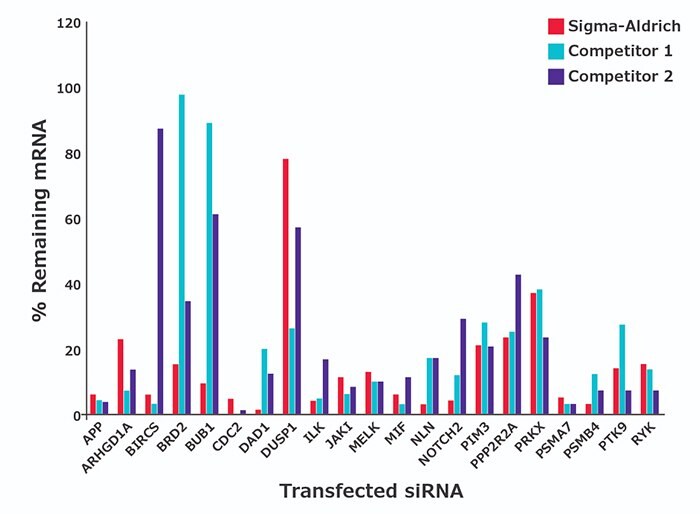
Figure 1.HeLa cells transfected with predesigned siRNA at a concentration of 30 nM. The percentage remaining gene expression levels were measured via qPCR 48 hours after transfection (relative to mock). Data represents the mean of four biological replicates.
| Validated siRNA by Gene Symbol | |||||||
|---|---|---|---|---|---|---|---|
| ABCC1 | CDC7 | DYRK1A | IGFBP4 | MLH1 | PIK3R4 | PRPS1 | SIK2 |
| ACP1 | CDC73 | DYRK1B | IL6ST | MSH2 | PIM2 | PRSS23 | SLC16A1 |
| ACP2 | CDK1 | EEF2K | ILK | MST4 | PIN1 | PSEN1 | SLC25A17 |
| ACVR1 | CDK11B | EIF2AK4 | IMPA2 | MTA2 | PINK1 | PSEN2 | SLC2A1 |
| ACVR1B | CDK17 | EIF4A3 | INPPL1 | MTM1 | PIP4K2B | PSMA7 | SLC30A1 |
| ADAM10 | CDK2 | EMR2 | IP6K1 | MTMR1 | PIP5K1A | PSMB4 | SLK |
| ADAM12 | CDK4 | ENPP1 | IP6K2 | MTMR2 | PKN2 | PSMB5 | SMU1 |
| ADAM15 | CDK5 | EPHA5 | IPMK | MTMR3 | PLAUR | PSMB6 | SNRK |
| ADIPOR1 | CDK6 | EPHB4 | IRAK1 | MTMR6 | PLK1 | PSPH | SORT1 |
| ADIPOR2 | CDK7 | F3 | IRAK4 | MTOR | PLK4 | PTK2 | SRC |
| ADRBK1 | CDK8 | FADD | ITPK1 | NADK | POLK | PTP4A1 | SRPK1 |
| AKT1 | CDK9 | FDFT1 | JAK1 | NCSTN | PPAP2C | PTPLA | ST7 |
| AKT2 | CDKL5 | FER | JUND | NDRG1 | PPAT | PTPN1 | STAT3 |
| ALPL | CDKN1B | FMNL1 | JUP | NEDD8 | PPM1D | PTPN11 | STK16 |
| APP | CDKN3 | FURIN | LANCL1 | NEK2 | PPME1 | PTPN12 | STK24 |
| ARAF | CELSR1 | FYN | LATS1 | NEK6 | PPP1CA | PTPN14 | STK3 |
| ARHGDIA | CERK | FZD4 | LDHA | NEK7 | PPP1CB | PTPN23 | STYX |
| ATF1 | CHEK1 | FZD5 | LEPR | NEK9 | PPP1CC | PTPN9 | TAOK1 |
| ATM | CHUK | FZD6 | LGALS3 | NET1 | PPP1R11 | PTPRE | TAOK3 |
| ATP6V0C | CKS2 | GLTSCR2 | LIMK2 | NF1 | PPP1R12A | PTPRF | TBK1 |
| ATR | CLPP | GPRC5A | LRP5 | NLN | PPP1R2 | PTPRJ | TEK |
| AURKB | CPE | GRB2 | LRP6 | NME1 | PPP1R3C | PTPRK | TESK1 |
| AXIN1 | CPT1A | GRK6 | LTBR | NME1-NME2 | PPP1R7 | PTPRS | TFRC |
| AXL | CRKL | GRN | LYN | NME2 | PPP2CA | RAB22A | TGFBR1 |
| BACE1 | CSK | GSK3B | MAD2L1 | NOTCH2 | PPP2CB | RAF1 | TGM1 |
| BACE2 | CSNK2A1 | HADHB | MANF | NPR1 | PPP2R1A | RAP1B | TGM2 |
| BAD | CTNNB1 | HBEGF | MAP2K2 | NR1H2 | PPP2R2A | RARG | THRA |
| BAG1 | CTSA | HCG 1757335 | MAP2K5 | NR1H3 | PPP2R5A | RB1 | TK1 |
| BCAT1 | CXCR4 | HDAC1 | MAP3K3 | NR2C2 | PPP2R5D | RELA | TKT |
| BIRC5 | CXCR7 | HDAC2 | MAP3K4 | NR2F2 | PPP2R5E | RHOA | TLR4 |
| BMP6 | CYLD | HECTD1 | MAPK14 | NUP85 | PPP3CC | RIOK3 | TM4SF1 |
| BMPR1A | DAD1 | HIPK2 | MAPK3 | OCRL | PPP5C | RIPK1 | TNFRSF10B |
| BRAF | DAPK3 | HIPK3 | MAPK6 | OXSR1 | PPP6C | RIPK2 | TRIM28 |
| BUB1 | DCN | HPRT1 | MAPK8 | PAK2 | PREPL | RNF10 | TWF1 |
| BUB1B | DDR1 | HSPA1A | MAPK9 | PASK | PRKACA | RNF5 | TYRO3 |
| CASK | DGKA | HSPA1B | MAPKAPK5 | PBK | PRKACB | ROCK1 | VEGFC |
| CCL2 | DMBT1 | HSPB8 | MARK3 | PCNA | PRKAG1 | ROCK2 | VIPR1 |
| CCNA2 | DNMT1 | HTRA1 | MASTL | PCSK9 | PRKAR1A | ROR2 | VRK2 |
| CCNC | DUSP10 | HUNK | MBTPS1 | PDE8A | PRKAR2A | RPS6KA3 | WNK1 |
| CCND1 | DUSP11 | ICAM1 | MELK | PDGFRB | PRKCA | RPS6KA4 | YES1 |
| CCT2 | DUSP12 | ICK | MET | PDK1 | PRKCI | RPS6KB1 | ZMPSTE24 |
| CD63 | DVL2 | IGF1R | MIF | PGK1 | PRKCZ | RYK | |
| CD82 | DVL3 | IGF2R | MINPP1 | PHPT1 | PRKDC | SHC1 | |
Select Citations
If additional help is needed, please consult our technical services group at oligotechserv@sial.com.
MISSION is a trademark of Merck KGaA, Darmstadt, Germany and/or its affiliates. Label License.
To continue reading please sign in or create an account.
Don't Have An Account?