R6265
EcoR I from Escherichia coli BS5
Restriction Enzyme
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
CAS Number:
EC Number:
MDL number:
UNSPSC Code:
12352204
NACRES:
NA.53
Recommended Products
grade
for molecular biology
Quality Level
form
buffered aqueous glycerol solution
concentration
10,000 units/mL
shipped in
wet ice
storage temp.
−20°C
Looking for similar products? Visit Product Comparison Guide
Specificity
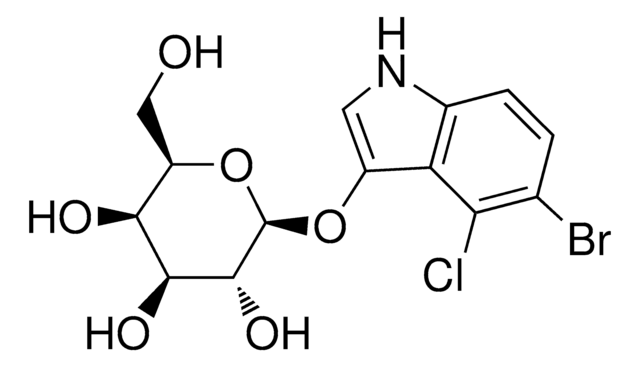
Recognition sequence: 5′-G/AATTC-3′
Cutting results: a 2-10-fold EcoR I overdigestion of 1 μg λ DNA substrate results in 100% cutting
Heat inactivation: 65 °C for 20 minutes.
Cutting results: a 2-10-fold EcoR I overdigestion of 1 μg λ DNA substrate results in 100% cutting
Heat inactivation: 65 °C for 20 minutes.
Application
EcoRI is a restriction endonuclease that is used in molecular biology applications to cleave DNA at the recognition site 5′-G/AATTC-3′, generating fragments with 5′-cohesive termini.
Other Notes
Supplied with 10x Restriction Enzyme Buffer SH (B3657).
Comment: Avoid suboptimal reaction conditions such as low salt, high pH (>8.0) and high glycerol (>5%) as they will alter EcoRI specificity and precipitate star activity.
Unit Definition
One unit is the enzyme activity that completely cleaves 1 mg of λDNA in 1 hour at 37 °C in a total volume of 50 ml of 1x digestion buffer SH for restriction enzymes. 1 mg pBR322 DNA is digested completely by 2 units of EcoR I.
Physical form
Solution in 10 mM Tris-HCl, pH 7.2 , 1 mM EDTA, 200 mM NaCl , 0.5 mM dithioerythritol, 50% glycerol (v/v), 0.2% Triton X- 100 (v/v), 4°C
related product
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Resp. Sens. 1
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
J Hedgpeth et al.
Proceedings of the National Academy of Sciences of the United States of America, 69(11), 3448-3452 (1972-11-01)
The sequence of DNA base pairs adjacent to the phosphodiester bonds cleaved by the RI restriction endonuclease in unmodified DNA from coliphage lambda has been determined. The 5'-terminal nucleotide labeled with (32)P and oligonucleotides up to the heptamer were analyzed
C Kessler et al.
Gene, 92(1-2), 1-248 (1990-08-16)
The properties and sources of all known class-I, class-II and class-III restriction endonucleases (ENases) and DNA modification methyltransferases (MTases) are listed and newly subclassified according to their sequence specificity. In addition, the enzymes are distinguished in a novel manner according
Cong Zhu et al.
Nucleic acids research, 41(4), 2455-2465 (2013-01-11)
Zinc-finger nucleases (ZFNs) have been used for genome engineering in a wide variety of organisms; however, it remains challenging to design effective ZFNs for many genomic sequences using publicly available zinc-finger modules. This limitation is in part because of potential
Annabel A Ferguson et al.
Methods in molecular biology (Clifton, N.J.), 940, 87-102 (2012-10-30)
The generation of transgenic animals is an essential part of research in Caenorhabditis elegans. One technique for the generation of these animals is biolistic bombardment involving the use of DNA-coated microparticles. To facilitate the identification of transgenic animals within a
TALENs and ZFNs are associated with different mutation signatures.
Yongsub Kim et al.
Nature methods, 10(3), 185-185 (2013-02-12)
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service