MetaPolyzyme, DNA Free Cell Lysis Enzymes for Microbiome Workflows
The downstream output for sample analyses intended for microbiome workflows are highly dependent on numerous factors during sample preparation. Key factors include the ability to efficiently lyse bacteria cells, extract their DNA and reduce reagent DNA contamination. Using our MetaPolyzyme, DNA free reagent in a microbiome workflow increases bacterial DNA extracted during cell lysis. Explore the benefits of MetaPolyzyme, DNA free reagents with sample data and methods provided below from our team of experts.
MetaPolyzyme, DNA free for DNA Extraction
The addition of MetaPolyzyme, DNA free enhances microbiome DNA extraction protocols and increases final DNA concentrations from microbiome samples. The addition of MetaPolyzyme, DNA free during DNA extraction also increases the number of taxa identified in microbial studies. MetaPolyzyme, DNA free is useful for low biomass samples for lysing cells without contributing to DNA background contamination. Explore two sample extraction studies below and discover the benefits of our MetaPolyzyme, DNA free reagent for your microbiome DNA extraction workflow needs. Saliva samples were used for the extraction studies. Here we show that the addition of MetaPolyzyme, DNA free to the DNA extraction workflow increases the recovery of microbial DNA and allows greater resolution of the microbial taxa.
Saliva DNA Extraction Sample Data Generated using MetaPolyzyme, DNA free
Saliva DNA extraction was performed on fresh, unfrozen saliva samples with the MagMAXTM Multi-Sample Ultra 2.0 Kit. Half of the sample aliquots underwent the protocol as described by the manufacturer, while the other half of the sample aliquots were pre-treated with MetaPolyzyme, DNA free. The results show that approximately twice as much DNA was extracted from samples pre-treated with MetaPolyzyme, DNA free compared to the manufacturer’s protocol (Figure 1). Furthermore, subsequent 16S amplicon library prep and sequencing revealed an increase in the number of microbial taxa identified (Figure 2). This data shows that the addition of MetaPolyzyme, DNA free enhances the lysis of microbial cells, improves the release of microbial DNA during DNA extraction, and is essential for sufficient data capture during a microbiome study.
DNA Extraction Sample Data from Saliva Using MetaPolyzyme, DNA free
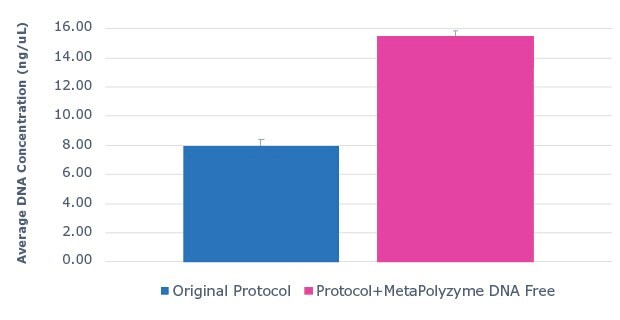
Figure 1:Use of MetaPolyzyme, DNA free enhances DNA extraction workflows. Average DNA concentration of sample extracted with MagMAX™ Multi-Sample Ultra 2.0 Kit from saliva with and without MetaPolyzyme, DNA free from DNA extraction shows approximately twice as much DNA is extracted with MetaPolyzyme DNA free treated samples.
Effect of MetaPolyzyme, DNA free on Taxa Identified in Microbial Populations
A.
B.
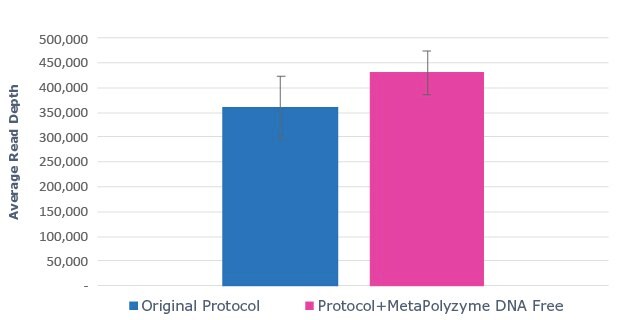
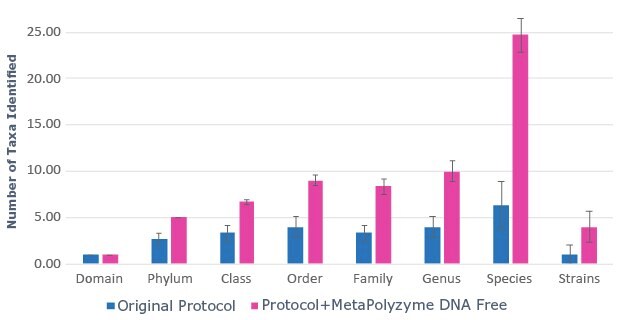
Figure 2: Use of MetaPolyzyme, DNA free increases the number of taxa identified in treated samples. A. Sequencing read depth for treated and untreated samples shows similar read depth. B. Taxa identified by bioinformatic analysis of treated and untreated samples shows increased number of taxa identified in samples treated with MetaPolyzyme, DNA free.
MetaPolyzyme, DNA free Sample Data Methods
DNA Extraction
Using MagMAXTM Multi-Sample Ultra 2.0 Kit for Saliva and KingFisher instrument using the MagMAXTM Ultra2 200uL FLEX Protocol in a 96 well plate format.
Bacterial Lysis with MetaPolyzyme, DNA free
200ul aliquots of saliva sample were combined with 4μL of MetaPolyzyme, DNA free (MAC4LDF) (lyophilized powder resuspended in 100μl PBS for final concentration of 1mg/mL) and incubated at 35°C for 30 min. Samples were mixed with 20μL Proteinase K and incubated at 56°C for 30 min. Samples were spun down at 10,000g for 10 min and supernatant was transferred. Samples were column purified with Sigma Aldrich’s GenEluteTM PCR Purification columns (Sigma NA1020), eluted in 50μL DNase and Microbial DNA free Trizma® Hydrochloride Elution Buffer (Sigma, SBR00051), and analysis proceeded using MagMaxTM DNA extraction.
Library Preparation
Libraries were prepared by targeted amplification of the variable V3-V4 regions of the bacterial 16S rRNA gene and attachment of Nextera XT indexes (Illumina Catalog #FC-131-2001) with PrimeStar® Taq DNA Polymerase (Takara, cat#R045A). Concentrations were measured using the Qubit dsDNA HS (Invitrogen, Cat# Q32851). Samples were pooled, then purified using Sigma Aldrich’s GenElute PCR Clean-Up Kit (Sigma NA1020), and KAPA Pure Beads (Roche 07983298001). Final library concentration determined by Invitrogen Qubit, and final size determined by Agilent DNA 1000 Kit (Agilent, #5067-1504) on the Agilent Bioanalyzer.
Sequencing
The library pools were sequenced on the Illumina® MiSeq™ system with a V2 reagent kit and MiSeq v2 500 cycle reagents by our Microbiome Multi-Omics Services.
Bioinformatics Analysis
Classifications of the microbial composition was performed using our Bioinformatics Database and MCamp platform.
To continue reading please sign in or create an account.
Don't Have An Account?