Epigenetics
Epigenetic Mechanisms
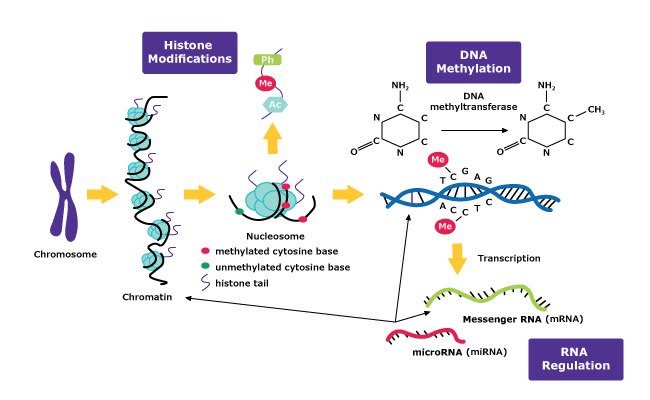
The field of epigenetics has become an essential area of focus for scientists working on cancer, neurodegenerative disease, and addiction research. Epigenetic mechanisms involve temporarily activating or repressing gene expression. Interestingly, these changes can be passed from generation to generation, even though they do not permanently change the DNA sequence. The three main mechanisms of epigenetics are DNA methylation, histone modification, and RNA regulation.
DNA Methylation
DNA methylation is the most well-known mechanism of epigenetics. It typically involves a methyltransferase enzyme that assists with the addition of a methyl group on the fifth position of cytosine (C5). This addition occurs mainly on cytosine-phosphate- guanine (CpG) dinucleotides. However, non-CpG methylation occurs as well. Analysis of DNA methylation is often performed to help understand gene expression. Examples of this type of analysis include methylation quantification by digestion of DNA with subsequent analysis through HPLC, mass spectrometry, or using sodium bisulfate conversion followed by PCR sequencing and analysis.
Histone Modification
Histone modification is another classic epigenetic mechanism. It involves various ways of altering histones by acetylation, methylation, phosphorylation, and other mechanisms that affect gene expression. Histones are proteins, that along with DNA, make up nucleosomes. Bundles of nucleosomes create the chromatin that make up chromosomes. In general, histone modifications take place at the histone N-terminal tails with high proportions of the amino acids lysine or arginine. One way to study this epigenetic regulation is through the use of chromatin immunoprecipitation (ChIP) assays.
RNA Regulation
Less is known about RNA regulation than the other epigenetic mechanisms. RNA signaling is thought to play a role in epigenetics through regulating chromatin structure. Researchers are investigating how mRNA and specifically non-coding RNA, such as long non-coding RNA, and micro RNA regulate gene expression. Additionally, Chromatin isolation by RNA purification (ChIRP) or RNA immunoprecipitation (RIP) assays can be used to understand the relationship between chromatin and RNA and the role RNA has in epigenetics.
Related Technical Articles
Related Protocols
- The DIG Gel Shift Kit uses recombinant terminal transferase and digoxigenin (DIG)-11- dideoxyuridine triphosphate (ddUTP), which makes the labeling reaction flexible.
- The Imprint® Methylated DNA Quantification Kit provides a high-throughput, non-radioactive means of measuring global DNA methylation shifts.
- Chromatin Immunoprecipitation qPCR for studying gene regulation across conditions.
- Chromatin Isolation by RNA purification (ChIRP) protocol for isolating RNA-bound genome regions.
- The Sigma Imprint Chromatin Immunoprecipitation Kit uses a plate based system to allow rapid ChIP assays in a high throughput format
- See All (5)
Find More Articles and Protocols
To continue reading please sign in or create an account.
Don't Have An Account?