Oligonucleotide Purification
During DNA manufacturing, each nucleotide is coupled sequentially to the growing chain via phosphoramidite chemistry. In each coupling cycle, a low percentage of the oligonucleotide chains do not extend, resulting in a mixture of full-length (n) and truncated (n-1, n-2, etc.) sequences (‘shortmers’). In addition, small-molecule impurities are the byproduct of the cleavage and deprotection process.
After cleavage, deprotection, and desalting (removes the small-molecule impurities), additional purification can separate the desired full-length sequence from undesired shortmers. The purity required for a specific technique / application (Table 1) depends on the potential problems caused by the presence of shortmers.
In addition, oligonucleotides that include common modifications, e.g. biotin, Amino C2 dT, etc. can be purified using any method described below. For oligonucleotides containing more complex dye structures, e.g. ROX™, TxRd (Sulforhodamine 101-X), etc., HPLC is the method of choice; only it can remove free dye that would otherwise interfere with performance of the intended technique / application.
Methods
Desalt
Every oligonucleotide is desalted, free of charge.
Desalting removes small-molecule impurities, e.g. acrylonitrile resulting from deprotection of the phosphodiester backbone, that are residual by-products from cleavage and deprotection. For many techniques / applications, including PCR, desalting is acceptable for oligonucleotides ≤35 bases as the overwhelming abundance of full-length sequence outweighs any contribution from shortmers (learn more about yield calculation). Oligonucleotides >35 bases may require additional purification, such as Reverse-Phase Cartridge (Cartridge).
No guaranteed purity level is provided with desalt-only purification since the process does not remove shortmers.
Cartridge
Separation on a reverse-phase cartridge offers the next level of purity (Figure 1). The basis of the separation is the difference in hydrophobicity between the full-length sequence (has hydrophobic 5’-DMT group) and shortmers (do not have 5’-DMT groups). While the full-length sequence is retained on the column, the shortmers are washed off. After cleaving the 5’-DMT on the cartridge, the expected full-length sequence is eluted and recovered. In addition, oligonucleotides modified with certain dyes at the 5’-end, e.g. Cyanine or WellRED, are compatible with cartridge purification due to the increased hydrophobicity imparted by the dye moiety.
As the oligonucleotide length increases, the proportion of shortmers containing a 5’-DMT group (the product generated by uncapped sequences) tends to increase. These undesired sequences will not be removed by cartridge purification and therefore, for longer oligonucleotides, high-performance liquid chromatography (HPLC) or polyacrylamide gel electrophoresis (PAGE) is recommended.
No guaranteed purity level is provided with cartridge purification. However, many years of experience has revealed that 65 - 80% full-length sequence (via analytical HPLC) is common.
RP-HPLC
Reverse-Phase High Performance Liquid Chromatography (RP-HPLC) operates on the same principle as a reverse-phase cartridge (Figure 1). However, the higher resolution allows for higher purity levels. RP-HPLC is an efficient purification method for oligonucleotides with dyes as their intrinsic hydrophobicity provides excellent separation of the desired sequence from those missing the dye, shortmers, and shortmers with deletions. Furthermore, RP-HPLC is the method of choice for large-scale synthesis due to the capacity and resolving properties of the column. However, resolution based on hydrophobicity decreases with the length of the oligonucleotide. Therefore, RP-HPLC is typically not recommended for purifying oligonucleotides >50 bases. Although longer oligonucleotides (up to 80 bases, in some cases longer) can be purified using this method, the purity and yields may be adversely affected.
Standard RP-HPLC typically achieves purity of >85% full-length sequence (via analytical HPLC). Higher purity levels may be achievable, depending on the nature of the sequence. For the exact nature of what RP-HPLC provides in terms of purity as well as any associated fees, please check with your local sales or customer service professional.

Figure 1.Separation via reverse phase: cartridge and HPLC. Both standard cartridge and RP-HPLC purification occur with the 5’-DMT on the sequence. 1) Upon entry into the column, the hydrophobic 5’-DMT of the desired sequence adsorbs onto the stationary-phase resin. 2) The shortmers, without 5’-DMT and therefore unable to interact with the resin, are washed off by the mobile phase. 3) The 5’-DMT is cleaved, and the desired sequence elutes.
IE-HPLC
Ion (specifically, anion)-Exchange High Performance Liquid Chromatography (IE-HPLC) is based on the number of charged groups (phosphate) in the oligonucleotide backbone. The anion-exchange method involves the use of a salt-gradient mobile phase on a quaternary ammonium stationary phase (Figure 2). The resolution is excellent for the purification of smaller quantities. However, IE-HPLC is limited by length, usually up to 40 bases. Longer oligonucleotides lead to lower resolution between the full-length sequence and shortmers; and, therefore, lower and less consistent purity.
The primary reason for using IE-HPLC as opposed to RP-HPLC is to purify oligonucleotides with significant secondary structure, typically found in sequences with high GC content. IE-HPLC is effective for such oligonucleotides because the mobile phase has a highly-alkaline pH, which disrupts hydrogen bonding, and therefore, secondary structure. This technique can be followed by RP-HPLC to add a second dimension to the separation process.
Please inquire for guaranteed percent full-length sequence.

Figure 2.Separation via IE-HPLC. IE-HPLC purification occurs without the 5’-DMT on the sequence. 1) Upon entry into the column, the polyanionic phosphates of the crude mixture form charge-charge interactions with the polycationic stationary-phase resin. 2) As the ionic strength of the mobile phase increases, the shortmers wash off. 3) The longer, desired sequence elutes last at an even higher ionic strength.
PAGE
Polyacrylamide Gel Electrophoresis (PAGE) uses a denaturing environment to separate oligonucleotides on the basis of molecular weight as opposed to charge (Figure 3). Under appropriate conditions, oligonucleotides differing by a single base can be resolved. This excellent size resolution typically results in the highest achievable purity level of any of the available purification methods. Minimum yields from PAGE are lower than from other methods due to the complex procedure required for extracting the oligonucleotides. This technique is recommended when high purity is required and for sequences ≥50 bases.
PAGE typically achieves purity of >95% full-length sequence. For the exact nature of what PAGE provides in terms of purity as well as any associated fees, please check with your local sales or customer service professional.
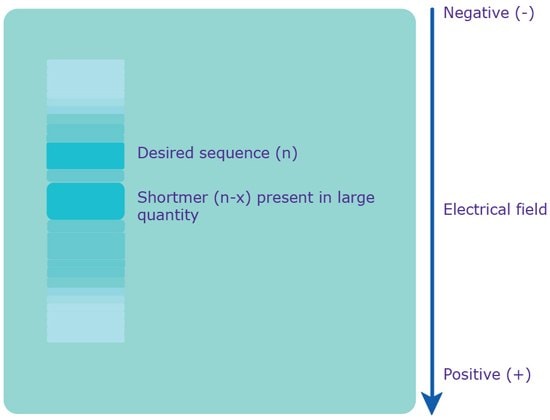
Figure 3.Separation via PAGE. An electrical field is applied to the gel, which causes the polyanionic oligonucleotide to travel to the positively charged anode. Since the charge is evenly distributed across the crude mixture, the various sequences are separated based on molecular weight, i.e. length. The shortmers travel faster, while the longer, desired sequence travels slower.
Gel Filtration
Gel filtration is recommended for smaller-scale oligonucleotides intended for in vivo experiments (Figure 4). Depending on manufacturing site, phosphorothioate oligonucleotides (S-Oligos), which are often used for in vivo antisense studies, may undergo gel filtration (please check with your local sales or customer service professional to see if this treatment is available in your geography). This purification removes trace amounts of synthesis, cleavage, and deprotection by-products (that were not removed by standard desalting), as well as purification solvents. These trace by-products, which are typically harmless in vitro, may lead to cytotoxic effects in vivo.
Additional in vivo options are available with our larger-quantity product, iScale Oligos™.

Figure 4.Separation via gel filtration. Gel filtration purification occurs without the 5’-DMT on the sequence. 1) Upon entry into the column, the mixture of the desired sequence with various by-product and solvent impurities traverse the stationary-phase resin. 2) As the mobile phase flows, the smaller impurities enter the gel pores, thereby slowing their progression through the matrix; the desired sequence does not enter the gel pores and therefore travels through the matrix to elute first. 3) The impurities finally pass through the gel pores and matrix; and are washed off.
Next-Gen Sequencing Oligos
The above traditional purification methods are effective at removing small-molecule impurities as well as increasing the percent full-length sequence to a desirable level. An additional type of impurity, cross contamination, has arisen as an issue due to continued growth in the use of extremely-sensitive techniques, such as next-generation sequencing (NGS). Traditional purification methods are not effective at removing cross-contaminating oligonucleotides.
Small amounts of cross contamination are a reality in any high-throughput oligonucleotide manufacturing facility. Typically, such small amounts have no discernable impact on traditional techniques / applications, such as PCR. However, most NGS platforms rely on index (barcode) adapters for multiplexing. If small amounts of the wrong barcode (due to cross contamination) are added to the wrong sequencing target, the problem is undetectable until the data analysis stage, making this problem costly in terms of time and money.
Next-Gen Sequencing Oligos (NGSO) have been developed to avoid this problem. NGSO is manufactured using proprietary purification methods and is recommended for any oligonucleotide intended to be used as an NGS adapter.
Conclusion
Oligonucleotides are best purified by different methods or combination of methods depending on the intended use. To start, consult Table 1 for the recommended methods based on technique / application. If additional help is needed, please consult our technical services group at oligotechserv@sial.com.
계속 읽으시려면 로그인하거나 계정을 생성하세요.
계정이 없으십니까?