Columns and Analytical Standards for Protein SEC
Judith Vajda (Tosoh Corp.), Eva Katharina Richter
Analytix 2014, Volume 1
Protein Size-exclusion Chromatography for Molecular Mass Determination

Aqueous size-exclusion chromatography (SEC) is well established for protein characterization. The technique has been used for quality control of biologics since the early days of recombinant protein development for pharmaceutical purposes, and method improvements have evolved. Today, (U)HPLC columns developed for specific separation tasks and ready-to-use standards facilitate method development and increase robustness of SEC methods.
Analytical size-exclusion chromatography can be used whenever a sample contains molecules that need to be separated and characterized based on size. The technique uses highly porous particles that are packed into a chromatography column. Depending on the size of a molecule in a sample, it may have access to all pores, limited access to some pores or no pore access. Based on this partitioning principle, the largest molecules elute first followed by sample components of progressively smaller sizes. Once a sample has been separated successfully, the sample molecules’ sizes can be determined using two basic detection methods. Static light scattering offers the opportunity for molecular mass determinations. Molecular masses are calculated of a light scattering signal and a refractive index signal and are independent of the hydration shell thickness of the molecules. The downside of this approach is the equipment cost and a comparably high vulnerability of the light scattering detector to background noise disturbance. Although light scattering has been established successfully for many applications, sometimes the more classical approach of molecular size determination outperforms static light scattering.
Size-exclusion Chromatography Calibration Curve
To produce a protein SEC calibration curve, a calibration mix consisting of various molecules of known size was injected onto the same column as used for the sample. The retention time of each component was obtained from the calibration chromatogram and plotted against the logarithmic molecular mass. Calibration curves were produced for three U(HPLC) columns that were developed for the analysis of immunoglobulin G (Figure 1). TSKgel® SuperSW mAb HTP was developed for easy method transfer from HPLC to fast UHPLC analysis, TSKgel® SuperSW mAb HR provides superior resolution for fragments, monomers, and aggregates in one run, and TSKgel® UltraSW Aggregate covers higher molecular weights and separates antibody dimers and higher aggregates best. Please find an up-to-date product list and ordering information at sigma-aldrich.com/tsk.
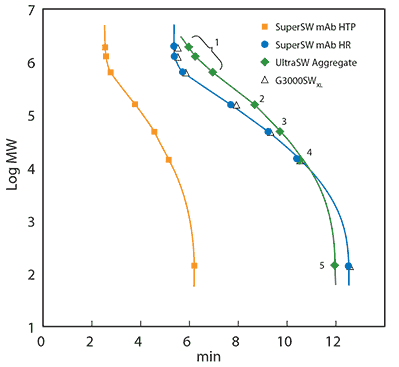
Figure 1. Calibration curves of TSKgel® SuperSW mAb HTP 4.6 mm ID x 15 cm L, TSKgel® SuperSW mAb HR, TSKgel® UltraSW Aggregate and TSKgel® G3000SWxl, both 7.8 mm ID x 30 cm L.
Mobile Phase: 0.2 M sodium phosphate buffer, pH 6.7 and 0.05% sodium azide. Calibrating proteins: thyroglobulin (640 kDa), γ-globulin (155 kDa), albumin (47 kDa), ribonuclease A (13.7 kDa) and ρ-aminobenzoic acid (0.14 kDa), all contained in the Protein Standard Mix 15-600 kDa. Injection volume: 10 µL. Flow rate: 0.35 mL/min (4.6 mm ID) or 1 mL/min (7.8 mm ID). Detection: UV @280 nm
The calibration curves show a linear correlation between the retention time and a certain range of molecular masses. This range of molecular masses is called the linear calibration range of a column and acts as the optimal working range for a separation. If a sample does not elute in this frame, the sample will usually require a SEC column with either smaller or larger pores. Also, proper column and eluent choice ensures that secondary interactions that may change the retention time are avoided.
Ready-to-Use Calibration Standard for Protein SEC
A calibration mixture should contain molecules that are similar to the analyzed sample molecules. Size-exclusion chromatography separates molecules according to their hydrodynamic size, which is dependent on the hydration shell thickness. The degree of molecular hydration in a given solvent is determined by molecular characteristics, such as the polarity. Often, polyethylene glycols are used as standards, because of their monodispersity. The molecules used for calibration should resemble just one molecular mass per peak. Also, polyethylene glycols are quite stable and can be stored at room temperature for years. Unfortunately, for protein analysis, polyethylene glycols have limited usefulness due to their much thicker hydration shell. For the molecular mass determination of proteins in SEC, proteins should serve as a calibration standard. Consequently, calibration standard handling becomes more elaborate, as typically protein-related problems such as protein degradation will emerge. Calibration mixtures based on proteins have limited shelf life, which means that either tiny amounts of standard need to be prepared, or large quantities of the calibration mix will be wasted. Overall, the preparation can be time-consuming. To avoid preparing standards, use ready-to-use standards.
The Protein Standard Mix for SEC offers full coverage of the molecular weight range from 15 kDa to 600 kDa. This qualifies it for the molecular mass determination of most proteins. The protein mix comes in 2 mL tubes that contain lyophilized protein powder and all necessary buffer ions. Simply add water and inject the calibration standard. This makes quick and easy column calibrations with a freshly prepared standard possible. When SEC is applied in the QC analysis of biopharmaceuticals the main purpose is quantification of sample components, and not determination of the molecular weights. For these types of SEC analysis, the injection of a protein standard solution is often used for system suitability tests. The protein standard is a convenient tool to facilitate the preparation of system suitability samples.
Column Calibration
When calibrating a column, it is important to apply the same conditions as used for analysis. Changing buffer systems changes the retention time considerably. For instance, arginine is a frequently-used additive for monoclonal antibody (mAb) aggregate analysis. It prevents secondary interactions of hydrophobic mAb aggregates and the stationary phase [1, 2 and 3] and shortens the retention time in SEC, which might also be due to a slightly altered hydration shell. Figure 2 illustrates the effect of an addition of 200 mM arginine to the liquid phase in SEC of an aggregated mAb sample.

Figure 2. Monoclonal antibody sample on TSKgel® UltraSW Aggregate with 0.1 M sodium-phosphate buffer containing 0.2 M arginine in the mobile phase (red). After 10 injections, the mobile phase was switched to sodium-phosphate buffer with 0.2 M sodium sulfate (gray). For both mobile phases, injection #10 is shown. Column: TSKgel® UltraSW Aggregate. Flow: 1 mL/min. Injection volume: 20 µL. Injected mass: 100 µg. Detection: UV @ 280 nm.
Retention shifts, such as for the arginine additive, would lead to miscalculating the molecular masses, if the calculation referred to a calibration standard separation that does not use arginine in the eluent. Similar effects have been observed for the change from phosphate buffer to HEPES (data not shown). Besides the applied liquid phase, the calibration standard concentration and injection volume are crucial for correct retention times and subsequent molecular mass calculation. Overloading SEC columns causes resolution loss and may lead to shifted retention times. Exemplary injection volumes and standard concentrations were injected onto TSKgel® SuperSW mAb HR. Sigma-Aldrich’s Protein Standard Mix was dissolved in 200 µL, 500 µL and 1000 µL water. Clearly, the highly concentrated calibration sample is not properly resolved. The effect is the most significant for the largest injection volume of 100 µL(Figure 3).

Figure 3.Protein Standard Mix 15-600 kDa on TSKgel® SuperSW mAb HR 7.8 mm ID x 30 cm L. Injection volumes are varied from 5 to 100 µL. The standard was prepared in different concentrations: 1X, 2X and 5X. Eluent: 200 mM sodium phosphate buffer, pH 6.7 and 200 mM sodium sulfate. Flow: 1 mL/min. Detection: UV @280 nm.
These results show the importance of a proper injection volume and concentration. Only then can reliable molecular masses be calculated. Moreover, permanently overloading the column may lead to early column deterioration. This is also true for the samples. The required amount of sample for a sufficient significant signal needs to be balanced against reduced column life time due to overloading. In general, 7.8 mm ID x 30 cm L columns achieve the best results and higher lifetimes when injecting 20 µL samples from 1 to 5 mg/mL.
Molecular mass determination using SEC can be an analysis with good accuracy and reproducibility. However, some basic methodical parameters need to be considered. Ready-to-use calibration standards allow removal of potential error sources, such as weighing low protein amounts. Easy handling and properly concentrated standards ensure the proper amount for column calibration or system suitability testing is injected.
References
TSKgel is a registered trademark of Tosoh Corporation
계속 읽으시려면 로그인하거나 계정을 생성하세요.
계정이 없으십니까?