Sample Preparation for Affinity Chromatography in Specific Groups of Biomolecules
Appendix 1, extracted from Affinity Chromatography Vol. 3: Specific Groups of Biomolecules, GE Healthcare, 2014
Samples for chromatographic purification should be clear and free from particulate matter. Simple steps to clarify a sample before beginning purification will avoid clogging the column, reduce the need for stringent washing procedures, and extend the life of the chromatographic medium.
Sample extraction procedures and the selection of buffers, additives, and detergents are determined largely by the source of the material, the stability of the target molecule, the chromatographic techniques that will be employed and the intended use of the product. These subjects are dealt with in general terms in the Strategies for Protein Purification Handbook and more specifically according to target molecule in the handbooks Affinity Chromatography, Vol. 1: Antibodies, 18103746 and Vol. 2: Tagged Proteins, 18114275 available from GE.
Sample stability
In the majority of cases, biological activity needs to be retained after purification. Retaining the activity of the target molecule is also an advantage when following the progress of the purification, since detection of the target molecule often relies on its biological activity. Denaturation of sample components often leads to precipitation or enhanced nonspecific adsorption, both of which will impair column function. Hence there are many advantages to checking the stability limits of the sample and working within these limits during purification.
Proteins generally contain a high degree of tertiary structure, kept together by van der Waals’ forces, ionic and hydrophobic interactions, and hydrogen bonding. Any conditions capable of destabilizing these forces can cause denaturation and/or precipitation. By contrast, peptides contain a low degree of tertiary structure. Their native state is dominated by secondary structures, stabilized mainly by hydrogen bonding. For this reason, peptides tolerate a much wider range of conditions than proteins. This basic difference in native structures is also reflected in that proteins are not easily renatured, while peptides often renature spontaneously.
It is advisable to perform some stability tests before beginning to develop a purification protocol. The list below shows examples of such testing:
- Test pH stability in steps of one pH unit between pH 2.0 and pH 9.0.
- Test salt stability with 0 to 2 M NaCl and 0 to 2 M (NH4)2SO4 in steps of 500 mM.
- Test the stability towards acetonitrile and methanol in 10% steps between 0% and 50%.
- Test the temperature stability in 10 °C steps from 4 °C to 40 °C.
- Test the stability and occurrence of proteolytic activity by leaving an aliquot of the sample at room temperature overnight. Centrifuge each sample and measure activity and UV absorbance at 280 nm in the supernatant.
Sample clarification
Centrifugation and filtration are standard laboratory techniques for sample clarification and
are used routinely when handling small samples.
It is highly recommended to centrifuge and filter samples immediately before chromatographic purification.
Centrifugation
Centrifugation removes lipids and particulate matter, such as cell debris. If the sample is still not clear after centrifugation, use filter paper or a 5 µM filter as a first step and one of the filters below as a second-step filter.
- For small sample volumes or proteins that adsorb to filters, centrifuge at 10 000 × g for 15 min.
- For cell lysates, centrifuge at 40 000 to 50 000 × g for 30 min.
- Serum samples can be filtered through glass wool after centrifugation to remove remaining lipids.
Filtration
Filtration removes particulate matter. Whatman™ syringe filters, which give the least amount of nonspecific binding of proteins, are composed of cellulose acetate (CA), regenerated cellulose (RA), or polyvinylidene fluoride (PVDF) (Table A1.1).
1The number indicates the diameter (mm) of the syringe filter.
For sample preparation before chromatography, select a filter pore size in relation to the bead size of the chromatographic medium (Table A1.2).
Check the recovery of the target protein in a test run. Some proteins adsorb nonspecifically to filter surfaces.
Desalting
Desalting columns are suitable for any sample volume and will rapidly remove LMW contaminants in a single step at the same time as transferring the sample into the correct buffer conditions. Centrifugation and/or filtration of the sample before desalting is still recommended. Detailed procedures for buffer exchange and desalting are given in the section Buffer exchange and desalting later in this appendix.
At laboratory scale, when samples are reasonably clean after filtration or centrifugation, the buffer exchange and desalting step can be avoided. For AC or HIC, it might be sufficient to adjust the pH of the sample. For IEX, it might be sufficient to dilute the sample to reduce the ionic strength.
Specific sample preparation steps
Specific sample preparation steps might be required if the crude sample is known to contain contamininants such as lipids, lipoproteins, or phenol red that might build up on a column. Gross impurities, such as bulk protein, should be removed before any chromatographic step.
Fractional precipitation
Fractional precipitation is occasionally used at laboratory scale to remove gross impurities from the sample. Precipitation techniques separate fractions by the principle of differential solubility. Because proteins differ in their degree of hydrophobicity, increased salt concentrations can enhance hydrophobic interactions between the proteins and cause precipitation. Fractional precipitation can be applied to remove gross impurities in three different ways, as shown in Figure A1.1.
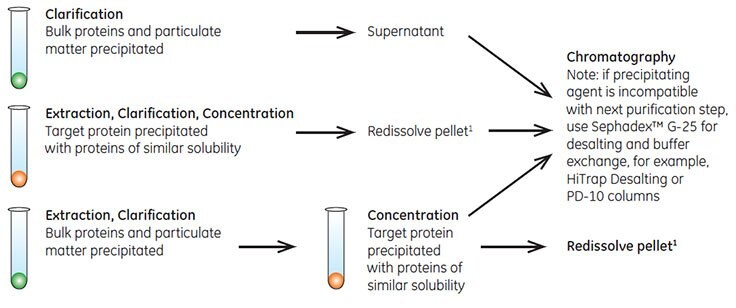
Figure A1.1.Three ways to use precipitation.
Precipitation techniques can be affected by temperature, pH, and sample concentration. These parameters should be controlled to ensure reproducible results.
Examples of precipitation agents are reviewed in Table A1.3. The most common precipitation method using ammonium sulfate is described in more detail.
Details taken from: Scopes R. K., Protein Purification, Principles and Practice, Springer, (1994), J. C. Janson and L. Rydén, Protein Purification, Principles, High Resolution Methods and Applications, second ed. Wiley Inc, (1998). Personal communications.
Ammonium sulfate precipitation
Some proteins can be damaged by ammonium sulfate. Take care when adding crystalline ammonium sulfate; high local concentrations can cause contamination of the precipitate with unwanted proteins.
For routine, reproducible purification, precipitation with ammonium sulfate should be avoided in favor of chromatography.
In general, precipitation is rarely effective for protein concentrations below 1 mg/mL.
Solutions needed for precipitation:
Saturated ammonium sulfate solution (add 100 g ammonium sulfate to 100 mL distilled water, stir to dissolve).
1 M Tris-HCl, pH 8.0.
Buffer for first purification step.
- Filter (0.45 µM) or centrifuge the sample (10 000 × g at 4 °C).
- Add 1 part 1 M Tris-HCl, pH 8.0 to 10 parts sample volume to maintain pH.
- Stir gently. Add ammonium sulfate solution, drop by drop. Add up to 50% saturation1. Stir for 1 h.
- Centrifuge 20 min at 10 000 × g.
- Remove supernatant. Wash the pellet twice by resuspension in an equal volume of ammonium sulfate solution of the same concentration (i.e. a solution that will not redissolve the precipitated protein or cause further precipitation). Centrifuge again.
- Dissolve pellet in a small volume of the buffer to be used for the next step.
- Ammonium sulfate is removed during clarification/buffer exchange steps with Sephadex G-25, using desalting columns (see Buffer exchange and desalting later in this appendix).
1 The percent saturation can be adjusted either to precipitate a target molecule or to precipitate contaminants.
The quantity of ammonium sulfate required to reach a given degree of saturation varies according to temperature. Table A1.4 shows the quantities required at 20 °C.
Resolubilization of protein precipitates
Many proteins are easily resolubilized in a small amount of the buffer to be used in the next chromatographic step. However, a denaturing agent may be required for less soluble proteins. Specific conditions will depend upon the specific protein. These agents must always be removed to allow complete refolding of the protein and to maximize recovery of mass and activity. A chromatographic step often removes a denaturant during purification. Table A1.5 gives examples of common denaturing agents.
Details taken from: Scopes R. K., Protein Purification, Principles and Practice, Springer, (1994), J. C. Janson and L. Rydén, Protein Purification, Principles, High Resolution Methods and Applications, second ed. Wiley Inc, (1998) and other sources.
Buffer exchange and desalting
Dialysis is frequently mentioned in the literature as a technique to remove salt or other small molecules and to exchange the buffer composition of a sample. However, dialysis is generally a very slow technique, requiring large volumes of buffer. There is also a risk of losing material during handling or as a result of proteolytic breakdown or nonspecific binding to the dialysis membranes. A simpler and much faster technique is to use a desalting column, packed with Sephadex G-25, to perform a group separation between HMW and LMW substances. Proteins are separated from salts and other small molecules.
In a fast, single step, the sample is desalted, transferred into a new buffer and LMW materials are removed.
Desalting columns are used not only to remove LMW contaminants, such as salt, but also for buffer exchange before or after different chromatographic steps and for the rapid removal of reagents to terminate a reaction.
Sample volumes up to 30% of the total volume of the desalting column can be processed. Sample concentration does not influence the separation as long as the concentration of proteins does not exceed 70 mg/mL when using normal aqueous buffers. The sample should be fully dissolved. Centrifuge or filter to remove particulate material.
For small sample volumes, it is possible to dilute the sample with the start buffer that is to be used for chromatographic purification, but cell debris and particulate matter must still be removed.
Volatile buffers such as 100 mM ammonium acetate or 100 mM ammonium hydrogen carbonate can be used if it is necessary to avoid the presence of NaCl.
Figure A1.2 shows a typical buffer exchange and desalting separation. The process can be monitored by following changes in UV absorption and conductivity.
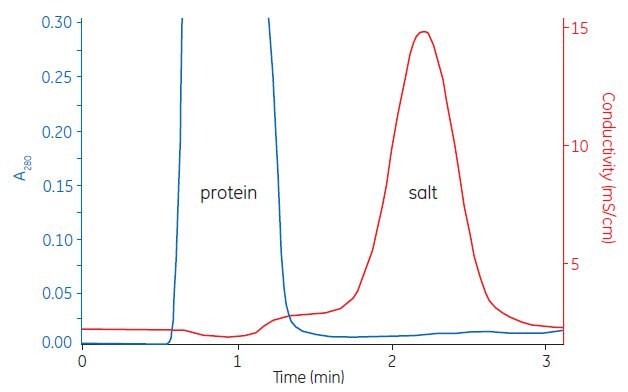
Figure A1.2. Buffer exchange of mouse plasma (10 mL) on HiPrep™ 26/10 Desalting.
To desalt larger sample volumes:
- Connect up to five HiTrap® Desalting 5 mL columns in series to increase the sample volume capacity, for example, two columns: sample volume 3 mL, five columns: sample volume 7.5 mL.
- Connect up to four HiPrep™ 26/10 Desalting columns in series to increase the sample volume capacity, for example two columns: sample volume 30 mL, four columns: sample volume 60 mL. Even with four columns in series, the sample can be processed in 20 to 30 min, at room temperature, in aqueous buffers.
Instructions are supplied with each column. Desalting and buffer exchange can take less than 5 min per sample with greater than 95% recovery for most proteins.
Manual desalting with HiTrap® Desalting 5 mL using a syringe
- Fill the syringe with buffer. Remove the stop plug. To avoid introducing air into the column, connect the column “drop to drop” to the syringe (via the adapter provided).
- Remove the snap-off end.
- Wash the column with 25 mL buffer at 5 mL/min to remove completely the 20% ethanol (supplied as storage buffer). If air is trapped in the column, wash with degassed buffer until the air disappears. Air bubbles introduced onto the column by accident during sample application do not influence the separation.
- Apply the sample (0.25 to 1.5 mL) using a 2 to 5 mL syringe at a flow rate between 1 to 10 mL/min. Discard the liquid eluted from the column.
- If the sample volume is less than 1.5 mL, change to buffer and proceed with the injection until a total of 1.5 mL has been eluted. Discard the eluted liquid.
- Elute the protein with the appropriate volume selected from Table A1.7 and collect the desalted protein.
Note: 5 mL/min corresponds to approximately 120 drops/min when using a HiTrap® 5 mL column. A simple peristaltic pump or a chromatography system can also be used for the desalting procedure.
The maximum recommended sample volume is 1.5 mL. Table A1.7 for the effect of reducing the sample volume applied to the column.
Removal of lipoproteins
Lipoproteins and other lipid material can rapidly clog chromatography columns and it is advisable to remove them before beginning purification. Precipitation agents such as dextran sulfate and polyvinylpyrrolidine, described under Fractional precipitation above, are recommended to remove high levels of lipoproteins from samples such as ascitic fluid.
Centrifuge samples when performing precipitation to avoid the risk of nonspecific binding of the target molecule to a filter.
Samples such as serum can be filtered through glass wool to remove remaining lipids.
Removal of phenol red
Phenol red is frequently used at laboratory scale as a pH indicator in cell culture. Although not directly interfering with purification, phenol red binds to certain purification media and should be removed as early as possible to avoid the risk of contamination. It is known to bind to AIEX media at pH > 7.0.
Use a desalting column to simultaneously remove phenol red (a low molecular weight molecule) and transfer sample to the correct buffer conditions for further purification, as described under Buffer exchange and desalting earlier in this appendix.
Removal of LMW contaminants
If samples contain a high level of LMW contaminants, use a desalting column before the first chromatographic purification step, as described under Buffer exchange and desalting earlier in this appendix.
To continue reading please sign in or create an account.
Don't Have An Account?