Optimizing Purification of Histidine-Tagged Proteins
Chapter 4, Extracted from Affinity Chromatography Vol. 2: Tagged Proteins, GE Healthcare, 2016
Three methods for optimizing purification of histidine-tagged proteins to achieve high purity are discussed in this chapter:
- Optimizing using imidazole
- Optimizing using different metal ions
- Optimizing using multistep purifications
For general purification of histidine-tagged proteins, including typical workflow, descriptions of available chromatography media and product formats, procedures, and troubleshooting hints, refer to Chapter 3.
Optimizing using imidazole
The presence of surface-exposed histidine residues or other complex-forming amino acids can lead to unwanted binding of untagged host cell proteins to purification media. These untagged proteins may elute with the target protein. The binding affinity of these contaminants is often lower than that of the tagged recombinant proteins; therefore, it may be possible to remove them by optimizing the separation conditions.
The examples below show how changes in imidazole concentration during binding affect the purity of the histidine-tagged target protein.
When using Ni Sepharose® excel and TALON® Superflow products it is usually not recommended to include imidazole during binding.
1. Imidazole as a competitive agent
Imidazole is utilized as a competitive agent for elution of histidine-tagged proteins. In addition, imidazole can be added in low concentrations in the sample and binding buffer in order to reduce the binding of contaminant proteins, and thus increase the final purity.
Histidine-tagged protein kinase G [(His)6-PknG] from Mycobacterium bovis was purified using a concentration of 45 mM imidazole in the sample and binding buffer. The medium used in the experiment was Ni Sepharose® High Performance (Chapter 3, Purification using Ni Sepharose® High Performance).
To achieve a higher protein concentration, the protein was eluted in a step gradient (Figure 4.1A). To demonstrate the advantageous effect of imidazole during binding, an additional purification was performed under the same conditions except that imidazole was omitted (Figure 4.1B). It is important to note that omission of imidazole is not generally recommended for Ni Sepharose® High Performance or Ni Sepharose® 6 Fast Flow; this example is provided solely to demonstrate the negative effect of its absence on the purity of the eluted target protein.
SDS-PAGE of the pooled elution fractions indicated a large improvement in purity of the target protein when 45 mM imidazole was included in the sample and binding buffer (Figure 4.1C). The yield of the target protein was in this case maintained in the sample with 45 mM imidazole present. Note that the optimal concentration of imidazole is protein dependent and thus must be determined case by case.



Figure 4.1. Purification of (His)6-PknG without (A) and with (B) 45 mM imidazole in the sample and binding buffer. For each chromatogram, the lysate of 2 l of E. coli culture (sample volume 26 mL; filtered through a 0.45 µm syringe filter) was loaded on a 2 mL Ni Sepharose® High Performance column (XK 16/20 column) using ÄKTApurifier chromatography system (now replaced by ÄKTA pure). The kinase was eluted in a two-step gradient with 50% and 100% of elution buffer. (C) SDS-PAGE (12% gel) of (His)6-PknG fractions showing eluates without (-) and with (+) 45 mM imidazole in the binding buffer. Data kindly provided by K. Hölscher, M. Richter-Roth and B. Felden de Neuman, GPC Biotech AG, Martinsried, Germany.
2. Determining optimal imidazole concentration using His SpinTrap
The imidazole concentration during binding and washing is an important factor for the final purity and yield of the target protein. His SpinTrap is a convenient and fast tool for determination of optimal imidazole concentration. Optimization is important for both purity and yield of the target protein. This was demonstrated by a series of experiments where a histidine-tagged protein, APB7-(His)6 (Mr 28 000), was purified on His SpinTrap using 5, 50, 100, or 200 mM imidazole in sample and binding buffers. The elution buffer contained 500 mM imidazole.
An imidazole concentration of 5 mM resulted in low purity of the eluted sample (Figure 4.2, lane 3), while an increase to 50 mM imidazole prevented binding of most contaminants and improved purity (Figure 4.2, lane 4). Including 100 mM imidazole in the sample and binding buffer lowered the yield while purity was improved marginally (Figure 4.2, lane 5). The lower yield can be explained by less binding of target protein due to the competitive effect of the high imidazole concentration during binding and washing. Further increase to 200 mM imidazole reduced yield even more (Figure 4.2, lane 6).
This example shows that higher imidazole concentrations during binding improve the purity, whereas too high of a concentration decreases the yield. The optimal imidazole concentration during binding is protein dependent. For many proteins, 20 to 40 mM imidazole is the best choice for His SpinTrap.

Figure 4.2.SDS-PAGE under reducing conditions (ExcelGel SDS Gradient 8–18) of histidine-tagged APB7 protein. The imidazole concentration during binding affects the final purity and yield (compare lanes 3, 4, 5, and 6).
Optimizing using different metal ions
The strength of binding between a protein and a metal ion is affected by several factors, including the structure and characteristics of the target protein, the presence and properties of the protein affinity tag, the properties of the metal ion, and the pH and composition of the binding buffer. As a result, Ni2+, the metal ion considered to have the strongest affinity to histidine-tagged proteins, may not always be the best choice for a given application. Under some circumstances, therefore, other transition metal ions, such as Co2+, Cu2+, Fe2+, and Zn2+, may be better suited.
In general, we recommend Ni Sepharose® High Performance or Ni Sepharose® 6 Fast Flow chromatography media, precharged with Ni2+ ions, for high capacity. If increased selectivity and higher purity would be advantageous, TALON® Superflow, precharged with Co2+, is a good choice. For altered selectivity, other metal ions can be tested using uncharged IMAC Sepharose® High Performance or IMAC Sepharose® 6 Fast Flow.
The following guidelines may assist in devising preliminary experiments to determine the metal ion most suitable for a given separation:
Ni2+ is generally used for histidine-tagged recombinant proteins.
Co2+ is also used for purification of histidine-tagged proteins, since it may allow weaker binding and reduce the amount of contaminants that may bind.
Cu2+ and Zn2+ are frequently used for purification of untagged proteins. Cu2+ gives relatively strong binding to a range of proteins; some proteins will only bind to Cu2+. Zn2+ ions often bind more weakly, a characteristic that is often exploited to achieve selective elution of the target protein. Both Cu2+ and Zn2+ can be used for histidine-tagged proteins and for process-scale separations.
Fe3+ is used more rarely than other metal ions. Take extra precautions when working with Fe3+, as it reduces easily in neutral solutions. Fe3+ is frequently used for purification of phosphopeptides, since the phosphate group has strong affinity for Fe3+. Chromatography on immobilized Fe3+ should be done at pH <3, to eliminate non-specific interactions from carboxyl groups. We also advise stripping immobilized Fe3+ ions after each run and recharging the column as required. Strongly bound Fe3+ ions and ferric compounds can be removed by leaving the medium in 50 mM EDTA overnight.
Below we present two examples showing how the selection of the most suitable metal ion and experimental conditions (including imidazole concentration) affects the purification of a given target protein.
1. Comparison study 2+, Zn2+, and Ni2+ on HiTrap IMAC FFusing Cu
In this study, APB7, a (histidine)6-tagged protein (Mr 28 000) expressed in E. coli BL-21, was purified on HiTrap IMAC FF 1 mL columns (prepacked with IMAC Sepharose® 6 Fast Flow) and charged separately with Cu2+, Zn2+, and Ni2+.
Screening experiments were performed to determine the optimal imidazole concentration for each ion [Figure 4.3 (A to C)]. The results of each of these three purifications indicate:
At 20 mM imidazole, there was significant leakage of target protein in the wash with Cu2+ and Zn2+ (arrows in Figure 4.3 A), a sign that imidazole concentration was too high to allow maximal yield. Very little leakage was seen with Ni2+. Purity was excellent in all three cases.
At 10 mM imidazole, leakage of target protein in the wash was significantly reduced for Cu2+ and Zn2+. The purity of the target protein in the eluted pool was similar with all three metal ions, but not as pure as with 20 mM imidazole.
At 5 mM imidazole, no leakage occurred with any of the metal ions. Ni2+ provided the purest protein, though still not as pure as with 20 mM imidazole.
Note that a large amount of sample was applied in the SDS-polyacrylamide gel. Because of this, the gels show a number of contaminants in the eluted material even though the purity was very high.
The results illustrate that, for any given metal ion, imidazole concentration can be adjusted to achieve high yield, high purity, or a successful compromise. IMAC Sepharose® media typically requires a slightly higher concentration of imidazole in the wash buffer than similar IMAC media on the market. A good starting point for most separations is to include 20 to 40 mM imidazole in the binding and wash buffers using IMAC Sepharose® 6 Fast Flow or IMAC Sepharose® High Performance. Be sure to use highly pure imidazole, which gives essentially no absorbance at 280 nm. To remove imidazole from the protein, use a desalting column (Chapter 11, Desalting/Buffer Exchange and Concentration).
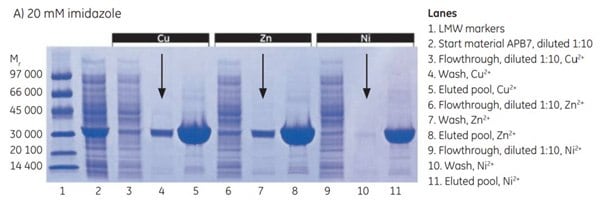


Figure 4.3.SDS-PAGE analyses (reducing conditions) of fractions from the purification of APB7 using IMAC Sepharose® 6 Fast Flow, prepacked in HiTrap IMAC FF 1 mL columns, charged with either Cu2+, Zn2+, or Ni2+, and with (A) 20 mM imidazole in the sample, (B) 10 mM imidazole in the sample, or (C) 5 mM imidazole in the sample. The gels were stained with Coomassie
2. Optimization o2+, Zn2+, Co2+, and Ni2+ on HiTrap IMAC HPf purity using Cu
Successful purifications require attention to the properties of the target protein and the affinity of the target protein toward the metal ion used. Four different HiTrap IMAC HP 1 m columns prepacked with IMAC Sepharose® High Performance were charged separately with four different metal ions: Cu2+, Zn2+, Co2+, and Ni2+. Purification performance was assessed using the target protein APB7 (Mr 28 000), expressed in E. coli.
The results demonstrate the importance of screening samples to determine the most suitable metal ion and purification conditions for specific target proteins. For APB7, the highest purity was achieved using Ni2+ or Co2+, but the difference compared with the results for Zn2+ and Cu2+ was small (Figure 4.4). The gradient elutions with imidazole employed in these examples also offer a methodology for selecting the appropriate imidazole concentration for a given purification.

Figure 4.4.Purification of APB7, a (histidine)6-tagged protein expressed in E. coli BL-21 on four different HiTrap IMAC HP 1 mL columns charged separately with metal ions. (A) Cu2+, (B) Zn2+, (C) Co2+, or (D) Ni2+. Pools selected after SDS-PAGE of individual 1 mL fractions (not shown) are indicated. (E) SDS-PAGE analysis: reducing conditions on ExcelGel SDS Gradient 8–18; Coomassie staining.
Optimizing using multistep purifications
Target protein can be further purified by adding one or more additional purification steps, as shown in the examples below. This topic is discussed in detail in Chapter 9 (Affinity chromatography in a purificationstrategy (CIPP)).
Below we present two examples showing successful multistep purification of the target proteins.
1. Two-step purification of 10-tagged protein using AC and SECa high-molecular-weight (histidine)
In a two-step purification of (His)10-trx-p450 (histidine-tagged at the N-terminus, 10 histidine residues) produced in E. coli, a HisTrap FF column was used in the first step. The eluted pool was then applied to a HiLoad 16/60 Superdex 200 pg column for further purification by SEC.
Three major bands were detected after the first purification step (Figure 4.5B, lane 5). Lane 6 (pool 2 from SEC) contains full-length target protein. Lanes 7 and 8 (pools 3 and 4, respectively) contain truncated forms of the target protein, as verified by N-terminal sequencing (data not shown). Lane 9 (pool 1) from SEC contains aggregated proteins. Thus subsequent SEC provided very good separation between the truncated forms and the full-length target protein, (His)10-trx-p450.

Figure 4.5.(A) Two-step purification of a high-molecular-weight (histidine)10-tagged protein using AC followed by SEC. (B) SDS-PAGE under reducing conditions and Coomassie staining.
2. Automatic three-step purification of unclarified cell lysate on ÄKTAxpress
An automated three-step protocol was used to purify histidine-tagged maltose binding protein from 100 mL of an unclarified E. coli cell lysate. The three steps were: Affinity chromatography (AC) using HisTrap FF crude (1 mL column), desalting (DS) using HiPrep 26/10 Desalting, and ion exchange chromatography (IEX) using Mono Q™ 5/50 GL. These are referred to as AC-DS-IEX in the image. As can be seen from the SDS-PAGE analysis, the target protein was obtained highly pure and in good yield.

Figure 4.6.(A) AC-DS-IEX with an enlargement of the IEX peaks and the collected pools to the right. Yield: 9.4 mg in pools 1 + 2. (B) SDS-PAGE of eluted pools from IEX. The gel was stained with Coomassie.
Per continuare a leggere, autenticati o crea un account.
Non hai un Account?