Genomic DNA Sample Purification and Quality Assessment
The availability of simple methods for purification of DNA and RNA has greatly facilitated the analysis and characterization of the genome and gene expression. There is a demand to isolate DNA and RNA rapidly and conveniently from a variety of cellular sources, including cells and tissues from mammalian, plant and bacterial cultures. Conventional approaches to DNA isolation and purification are based on multi-step procedures involving phenol/chloroform, anion exchange, or silica gel exchange systems. Purification of RNA typically involves chaotropic salt and phenol/chloroform with a further oligo-dT purification step if mRNA is required.
The quality of the template material that is included into a PCR or RT reaction has a profound effect upon the reliability of the resulting data1. It is critical that the quality of the target material is determined and reported2 and it is preferable to use the highest quality nucleic acid possible. This chapter contains an in depth analysis of the effect of target quality on resulting qPCR and RT-qPCR data with consideration of both purity and integrity.
Nucleic Acid Purification
Nucleic acid purification is the first requirement for most experiments utilizing molecular biology. DNA and RNA samples are often obtained via “crude” preparations. These purification methods can be rapid and direct and are especially useful when samples are being used in PCR/RT-PCR where only a small amount of DNA or RNA is required. Some applications require a sample that is purified via conventional methods; the final application will dictate the best method to select. When the nucleic acid from biological material needs to be intact, pure, concentrated and without inhibitors of downstream enzymatic reactions, the extraction procedures require addition of chaotrophic agents.
Chaotropic Agents
A chaotropic agent is a substance which denatures proteins, DNA, or RNA by disrupting the three dimensional structure of the molecule. Chaotropic agents interfere with stabilizing intra-molecular interactions that are mediated by noncovalent forces such as hydrogen bonds, van der Waals forces and hydrophobic effects. Commonly used chaotropic agents include urea (6–8 M), guanidine HCL (6 M) and lithium perchlorate (4.5 M). Guanidine thiocyanate is a potent protein denaturant that is stronger than guanidine HCl and is often used during the isolation of intact ribonucleic acid to eliminate RNase activity. Many RNA isolation protocols include the addition of mercaptoethanol, in addition to a chaotrope, to provide a reducing environment that denatures the four disulfide bridges formed between the eight cysteine residues of Ribonuclease A (RNase A). The combination of a chaotropic agent and a reducing agent such as mercaptoethanol causes the ribonuclease to unfold and completely lose its activity.
Extraction of Genomic DNA
Crude Preparation
The Extract-N-Amp™ kits can be used to extract PCR-ready genomic DNA (gDNA), rapidly and efficiently, from almost any sample type, using a simple, one-tube protocol that takes 15 minutes or less, depending on the sample. The Extract-N-Amp Kits have been used to extract gDNA from samples such as whole blood, hair follicles, mouse tails snips, tissue-culture cells, buccal swab cells, Drosophila melanogaster, various plants and seeds. Unlike full purification procedures, this approach avoids mechanical disruption, organic extraction, column purification, or precipitation of the DNA. The kits contain reagents for extracting and amplifying DNA which may be used in many downstream applications, including PCR/qPCR analysis.
Purification of Genomic DNA
DNA can be isolated from almost any cellular or tissue source. Purification of gDNA requires removing it from the cell while protecting against degradation. Isolation procedures must also be gentle enough to protect the long DNA strands from mechanical stress. The sample is first harvested in a buffer such as phosphate buffered saline (PBS), usually containing a detergent such as Triton™ X-100 or Sodium dodecyl sulphate (SDS) to lyse the cells and solubilize proteins and lipids. Proteinase K is added to separate clumped cells and tissues and inactivate enzymes by proteolytic digestion. Ethylenediaminetetraacetic acid (EDTA) is a chelator that acts as a scavenger for metal ions in solution and is added to DNA purification buffers to remove magnesium ions (Mg2+). Mg2+ is an essential co-factor for DNases, therefore removal of Mg2+ inactivates DNases, preventing DNA degradation. RNase is often included to degrade the RNA present in the cells.
After lysing the cells and degrading the RNA, proteins and lipids, the DNA must be separated from the remaining remnants of these materials. The classical purification methods use phenol/chloroform3 to remove the proteins, leaving DNA and other water-soluble materials behind. TRI Reagent® uses an adaptation of this method. Alternatively using systems such as the GenElute™ gDNA purification kits, DNA is adsorbed onto silica-based membranes (in single column and 96-well formats), in the presence of chaotropic salts, which disrupt protein structure. When purifying DNA from plant material, the tissue must first be mechanically disrupted by grinding in liquid nitrogen, whereas whole blood or bacteria are pre-incubated with enzymes to ensure efficient cell lysis and DNA release. The DNA is washed while bound to the membranes and then eluted by changing the salt concentration and is subsequently released with detergent and a chaotropic agent. Proteins, polysaccharides and cell debris are eliminated with a precipitation procedure followed by centrifugation through a filtration column.
Purification of Total RNA
Purification of cellular RNA is required for studies of gene expression or other cellular functions that are regulated by RNA. Total RNA is the RNA that is transcribed from cellular DNA (gDNA and mitochondrial DNA, mtDNA) and generally refers to a sample containing: Ribosomal, transfer and messenger RNA (rRNA, tRNA and mRNA) and does not include microRNA (miRNA) or smaller non-coding RNAs (ncRNA). Total RNA is the fraction of interest which is isolated for gene expression analysis.
The isolation of RNA is challenging because of the ubiquitous presence of ribonuclease enzymes (RNases) in cells and tissues, which rapidly degrade RNA. Therefore, RNase inhibitors are included during the purification process. The most common method for purification is guanidinium thiocyanatephenol/chloroform extraction. This is the principle behind TRI Reagent®, which is a monophase solution containing phenol and guanidine thiocyanate. The tissue or cell sample is homogenized or disrupted in the TRI Reagent, chloroform is mixed with the lysate and then the mixture is separated into three phases by centrifugation. The aqueous phase containing the RNA is removed and the RNA is precipitated using isopropanol. GenElute RNA isolation kits capture the RNA onto silica resin following cell lysis. GenElute kits use guanidine thiocyanate and 2-mercaptoethanol to inactivate RNases when isolating total nuclear and cytoplasmic RNA. In the presence of guanidine thiocyanate, proteins dissolve readily, cellular structures disintegrate and nucleoproteins dissociate from nucleic acids as protein secondary structure is lost. Furthermore, since guanidine thiocyanate is more potent than guanidine HCL, RNases are permanently denatured. Lysates are spun through a filtration column to remove cellular debris and shear DNA. The filtrate is then applied to a high capacity silica column to bind total RNA, followed by washing and elution with RNase free water or buffer.
Of the total RNA sample, the majority is rRNA (~80%) while the mRNA fraction is only 2–5%. For some procedures, it may be desirable to purify the mRNA from the vastly more abundant rRNA and tRNA. The GenElute mRNA Kits provide convenient procedures for isolating polyadenylated mRNA from previously prepared total RNA or directly from mammalian cells and tissues. For direct mRNA preparation, cells or tissues are disrupted with SDS/proteinase K digestion to release RNA and eliminate RNases. Oligo dT30 covalently linked to 1 μm polystyrene beads is then used to capture polyadenylated mRNA by hybridization. The polystyrene beads remain suspended during hybridization, eliminating the need for mixing or rocking. The mRNA-bead complexes are washed on a microcentrifuge spin filter and then the purified mRNA is eluted.
Many of the extraction methods used for purification of “total RNA” specifically select larger molecules and so are not suitable for isolation of smaller ncRNAs, including miRNAs. There are specifically designed kits for purification of fractions of smaller RNA species, which may be selected when these are the template of interest.
Purification of Micro RNA (miRNA) and Non-coding RNA (ncRNA)
Purification of miRNA and other, smaller ncRNA molecules is more challenging due to their short lengths. It is important to select an appropriate RNA isolation method that retains small RNA and avoid using a total RNA purification kit unless the manufacturer specifically states that total RNA includes small RNAs <100 bases. Extraction methods for ncRNA target differences between RNA molecules and any other contaminating factors, using specific column binding or precipitation. RNAzol® is an acidic guanidine thiocyanate-phenol mixture that disrupts cells and tissues and, with the addition of chloroform or bromochloropropane, separates all RNA species from proteins and DNA. Purification methods then allow separate preparation of small RNA, mRNA or total RNA samples such that each of these fractions can be analyzed in parallel in downstream applications. The miRPremier® microRNA Isolation kits are silica-based bind-wash-elute kits specifically designed for small RNA recovery. The miRPremier kit uses a salting out approach to remove larger molecules before binding small RNA to the column, because the amount of ethanol needed to bind small RNA to silica (silicon dioxide) will also precipitate proteins and other macromolecules, creating a viscous suspension that can clog columns and prevent efficient recovery of RNA. An alternative approach to circumvent this problem is to use a pre-clearing step to remove macromolecules, such as by extracting with RNAzol or a pre-binding column, so that sufficient alcohol can be added to bind small RNA without clogging the RNA binding column.
Nucleic Acid Sample Quality Assessment
Sample handling and nucleic acid extraction procedures are variable, therefore it is critical to define a reliable protocol for analysis of sample quality and quantity and to include details of this analysis in publications2. The quantity of RNA is largely determined by the yield of rRNA, whereas the quality of RNA is determined by the purity (absence of inhibitors) and the integrity of the RNA molecules. However, when measuring the quality of a sample the purity and integrity influence the results; it is difficult to determine the purity and integrity of a sample of low concentration and the presence of impurities can interfere with quantification. There are a number of techniques that may be used to assess nucleic acid quantity and quality but none of these, alone, provide all of the information required to describe a sample fully. Therefore, it is important to consider a suite of options to ensure that sample quality will not influence the final analysis data1.
Nucleic Acid Quantification
UV Spectroscopy
Nucleic acids have traditionally been quantified by measuring UV absorption using a spectrophotometer. The absorbance of a diluted sample of DNA or RNA is read at 260 nm and 280 nm. The Optical Density (OD) at 260 nm is used to determine the RNA or DNA concentration in a solution using the Beer-Lambert law, which predicts a linear correlation between absorbance and concentration.
Beer-Lambert Law
A = εbc
A = absorbance
b = path length of the cuvette in cm (typically 1 cm)
c = analyte concentration
ε = extinction coefficient
For RNA, ε = 40 μg/mL
For DNA, ε = 50 μg/mL
An OD at 260 nm (A260) reading of 1.0 is equivalent to approximately 40 μg/mL of pure RNA and 50 μg/mL of pure double-stranded DNA. However, the Beer-Lambert law is only valid for OD readings up to 2 and the stated OD/concentration relationship relies upon the samples being pure. Evidently, contaminating substances with absorption at 260 nm or 280 nm will affect the estimated OD reading. Pure RNA has an A260/A280 of 2.1, whereas pure DNA will have an A260/A280 of 1.8.
The OD of potentially contaminating substances such as proteins, chaotrophic salts and phenol can also be determined if absorbance of the sample is measured at 280 nm and 230 nm (A280 and A230, respectively). In this way, a ratio of A260/A280 and A260/A230 may be used as an indicator of sample purity. However, it should be noted that the determination of protein contamination in DNA is very insensitive when using this approach4.
Additionally, pH and the ionic strength of the buffer also disturb the OD reading. It has been shown that there is significant variability in the A260/A280 ratio when different sources of water are used to perform the spectrophotometric determinations. For example, variation in the pH of water between pH 5.4 to 7.5–8.5 used for suspending RNA significantly increased RNA A260/A280 ratios from approximately 1.5 to 2.05.
It is important that both the A260/A280 ratio and the A260/A230 ratios are close to 2.0 (Table 7.1).
TRIS, Ethanol and Isopropanol Contamination
Effect of TRIS contamination: TRIS contamination does not influence the absorbance spectrum of RNA/DNA significantly. Effect of ethanol contamination: Ethanol does not have a significant effect on the absorbance spectrum of RNA/DNA when measured in TE pH 8.0. However, when the measurement is made in pure water, the A260/A280 ratio may be reduced and so a ratio lower than 2.0 may indicate ethanol contamination.
Effect of ethanol contamination: Ethanol does not have a significant effect on the absorbance spectrum of RNA/ DNA when measured in TE pH 8.0. However, when the measurement is made in pure water, the A260/A280 ratio may be reduced and so a ratio lower than 2.0 may indicate ethanol contamination.
Effect of isopropanol contamination: Isopropanol does not have a significant effect on the absorbance spectrum of RNA/DNA when measured in TE pH 8.0. However, when the measurement is made in pure water, the A260/A280 ratio can be reduced and so a ratio lower than 2.0 may indicate isopropanol contamination.
Protein, Guanidine Isothiocyanate and Phenol Contamination
Effect of protein contamination: A sample containing a contamination of 0.01% BSA can show an almost normal absorbance spectrum, although the A260/A280 ratio may fall below 1.9 (RNA) or 1.7 (DNA), which is a warning that something is contaminating the sample. At 0.5% BSA, the absorbance spectra appear abnormal and this is a clear indication that there is a significant level of contamination in the sample. Therefore, high concentrations of protein may be detected by abnormal A260/A280 ratios but low and moderate amounts are not.
Effect of guanidine isothiocyanate contamination: Guanidine isothiocyanate has little influence on the A260/A280 ratio, therefore, guanidine isothiocyanate has a small effect on the quantification of RNA. However, the presence of guanidine isothiocyanate has a very strong effect on the A260/A230 ratio; at a 0.5% contamination, the A260/A230 ratio drops below 0.5. Samples with suspected guanidine contamination should be avoided since this would have a deleterious effect on downstream enzymatic reactions.
Effect of phenol contamination: Phenol has a very strong effect on the quantification of RNA. At 0.5% phenol contamination of a RNA sample at 50 ng/μL, the measured concentration is over three times higher. This strong disturbing effect of phenol on the quantification of RNA is due to the contaminating absorption peak at 270 nm. At very low RNA concentrations, below 10 ng/μL, this contaminating peak is often confused for RNA. However, RNA absorbance is always at 260 nm and never at 270 nm, therefore if the peak is at 270 nm, it is due to a contamination. After RNA has been isolated using a method based on phenol, the erroneous overestimation of RNA concentration is a serious problem. This is particularly the case when the RNA is of low concentration.
Automated UV Spectrophotometry
While conventional UV spectrophoresis required measurements to be carried using relatively high volumes of sample, there are now systems such as the NanoDrop® 2000 UV-Vis Spectrophotometer that are automated and support highly accurate analyses of extremely small sample sizes. The sample retention systems eliminate the need for cuvettes and capillaries, which decreases the amount of sample analyzed to between 0.5 μL and 2 μL. The NanoDrop provides a scan of the absorbance from about 200 nm to 350 nm, which is the relevant region for determining RNA/DNA concentration and purity.
Fluorescence-based Nucleic Acid Quantification Systems
The use of fluorescent dyes to quantify nucleic acids is an alternative to absorbance spectrophotometry6,7. Determination of nucleic acid concentration using fluorescent methods utilizes the binding of small molecules or dyes to nucleic acids and measures the subsequent changes in fluorescence characteristics. Although more expensive than absorbance spectrophotometry, fluorescence-based quantification is more sensitive, precise and may be specific for the nucleic acid of interest.
Since fluorometers measure fluorescence in relative rather than absolute units, the measurement is first calibrated with a known concentration of a standard nucleic acid solution with characteristics similar to the sample to be measured. Following calibration, a single measurement can establish the concentration of nucleic acid in the solution, but typically a standard curve will be required to ascertain the linearity of the assay in the range measured.
Automated systems such as the QuBit® 2.0 fluorometer (Life Technologies) can be used with a range of different fluorescence based quantification assays for the measurement of nucleic acid concentration in solution. The assays demonstrate a wide dynamic range for detection and are capable of accurately analyzing small samples.
It is critical to observe that the OD reading is a measure of absorption and cannot be used as a complete evaluation of sample quality. Similarly fluorescent-based systems provide a measure of quantity and not quality. For example, these cannot be used as a test for sample integrity and a suitable analysis must also be performed, such as a capillary electrophoresis, gel resolution, or the 3’/5’ ratio assay (as described below).
Gel Electrophoresis and Capillary Nucleic Acid Quality Analysis Systems
Gel Electrophoresis
RNA, although thermodynamically stable, is rapidly digested in the presence of almost ubiquitous RNase enzymes. As a result, shorter fragments of RNA commonly occur in a sample, which can potentially compromise the results of downstream applications. The degradation process of RNA is only partly understood and is dependent on the type of RNase that is present. Also, the quality of RNA from a given extraction can vary extensively and needs to be under constant surveillance. Therefore, it is important to determine the integrity of the RNA samples.
In order to evaluate potential degradation, electrophoretic methods have been applied that separate the samples according to the size of the comprised molecules. Historically, RNA integrity has been evaluated using agarose gel electrophoresis stained with ethidium bromide, which produces a defined banding pattern. Total RNA run on a denaturing agarose gel displays two distinct ribosomal fragments corresponding to either 18S or 28S for eukaryotic RNA or 16S and 23S for prokaryotic RNA and additional fragments of smaller RNA species. Traditionally, the intensity of these rRNA bands on denaturing agarose gels have been used to calculate a ratio that served as an indication of RNA integrity. RNA is considered to be of high quality when the ratio of eukaryotic 28S:18S band intensity is about 2.0.
Figure 1 shows an example of total RNA evaluation by ethidium bromide-stained agarose gel electrophoresis. For
good quality total RNA, the two largest rRNAs appear as discrete bands at approximately 5 kb and 2 kb and the upper band should have approximately twice the intensity of the lower. The mRNA may appear as a light smear or may barely be evident, mostly between the two rRNA bands.

Figure 1.Evaluation of RNA integrity by agarose gel analysis. Samples of total RNA (2 μg) were fractionated on a 1% agarose gel in TBE buffer and stained with ethidium bromide. Lanes 1 and 2 show RNA of high quality while lanes 3 and 4 show degraded RNA.
However, gel electrophoresis is an insensitive method for determination of sample integrity and requires large concentrations of sample. When working with small samples there is rarely a sufficient amount to run gel electrophoresis and also perform all desired experiments. Since this approach relies on human interpretation of gel images, it is subjective, hardly comparable from one lab to another and the resulting data cannot be processed digitally.
Some of these issues are addressed by the analysis of total RNA by automated capillary electrophoresis.
Capillary Nucleic Acid Quality Analysis Systems
Nucleic acid samples can be analyzed and compared using instrumentation such as the Agilent 2100 BioAnalyzer or Bio-Rad Experion. Using these systems, electrophoretic traces are used to assign numerical integrity values or integrity categories.
These instruments use a lab-on-a-chip approach, combining capillary electrophoresis with fluorescent detection. The electrophoretic process carried out on the chip is based on traditional gel electrophoresis principles that have been miniaturized, which reduces sample consumption and separation time.
The chip accommodates wells for samples, gel and an external standard (fragment size ladder). During manufacturing, microchannels are fabricated in glass to create interconnected networks amongst the wells. These micro-channels are then filled with a sieving polymer and fluorescence dye. Electrodes are inserted in the wells and the chip becomes an integrated electrical circuit.
Charged bio-molecules such as DNA or RNA are driven through the matrix in response to a voltage gradient. Due to a constant mass-to-charge ratio and the presence of a sieving polymer matrix, the molecules are separated by size such that smaller fragments migrate faster than larger ones.
Dye molecules intercalate into nucleic acid strands and these complexes are detected by laser-induced fluorescence. Data is then translated into electronic gel-like images and electropherograms.
The Agilent 2100 BioAnalyzer is used in conjunction with the RNA 6000 Nano and the RNA 6000 Pico LabChip kits. This bio-analytical device is based on a combination of microfluidic chips, voltage-induced size separation in gel filled channels and laser-induced fluorescence (LIF) detection on a miniaturized scale. Twelve samples can be processed sequentially while consuming very small amounts of each sample. RNA molecules are stained with an intercalating dye and detected by means of LIF. Data are archived automatically and available as electropherograms, gel-like images and in tabular format.
The BioAnalyzer has a good linear dynamic range and the RNA 6000 Nano assay can be used to analyze RNA samples of concentrations ranging from 25 ng/μL to 500 ng/μL. Although the lower quantitative limit of the RNA 6000 Nano assay is specified as 25 ng/μL, it is recommended to use at least 50 ng/μL for a meaningful RNA Integrity Number (RIN) calculation. When using lower concentrations, higher sample to sample variances of the RIN may be observed. The Pico assay can be used to determine RNA integrity for samples of concentrations in the range 50 pg/μL to 5 ng/μL.
The Agilent 2100 BioAnalyzer software is used to evaluate the proportion of RNA detected before, between and after the rRNA peaks in order to determine a relative integrity number (RIN) for the RNA sample. Intact RNA has a RIN of 10, whereas completely degraded RNA has a RIN of 1. In this way, interpretation of an electropherogram is facilitated and comparison of samples is possible.
Partially degraded RNA (RIN <9) may yield satisfactory results in RT-qPCR but it is likely that this depends upon the targets analyzed, degree of sensitivity required and the RT strategy. A RT-qPCR strategy that uses two steps with oligo-dT to prime reverse transcription and PCR primers near the 5’-end of a long cDNA will require much higher integrity samples than a strategy that uses one-step RT-qPCR with gene-specific primers. The effect of different RT strategies and their tolerance of degraded templates is discussed in detail (see Reverse Transcription). In addition, higher sample integrity will be required to detect a rare rather than an abundant mRNA target. Therefore, the effect of template degradation on the ratio of targets of different abundance (e.g., test and reference genes) should also be considered. The correlation between RIN and RT-qPCR success should be determined empirically for each assay. Although the RIN tremendously facilitates the assessment of the integrity of RNA preparations and makes it much easier to compare samples or RNA isolation procedures, it is not yet possible to determine in advance whether the RNA may be suitable for a given experiment. Figure 2 shows the Agilent BioAnalyzer traces for two RNA samples. No difference was detected in the yield of several rare mRNA targets in cDNA generated with one-step RT-qPCR using gene-specific primers. However, the same mRNA targets were detected up to 2 cycles later, indicating 4-fold less mRNA, in the lower integrity sample (11, RIN = 7.0) than in the higher quality sample (9, RIN = 9.8) when using two-step RT-qPCR with oligo-dT to prime the RT.
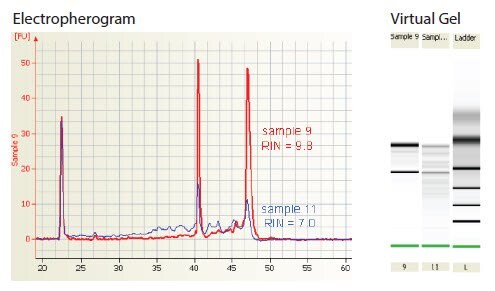
Figure 2Electropherogram showing the evaluation of RNA integrity using the Agilent 2100 BioAnalyzer. Samples of total RNA preparations (~150 ng) were fractionated on a RNA 6000 Nanochip and resolved using the Agilent 2100. Integrity was evaluated and ascribed a RIN
Sample quality is critical and therefore must be verified before the samples are used in qPCR assays. It has been clearly demonstrated that analysis of degraded RNA samples may result in poor quality data1, although this is not always the case8 and is partly dependent on RT protocol (Reverse Transcription)9.
Although the capillary systems are a great improvement over conventional gel electrophoresis, they are specialized pieces of equipment, expensive and are not amenable to high throughput. In addition, it has been observed that the RIN may not be the panacea measurement that would be desired. Figure 3 shows samples which have been subjected to incubation on bare human skin for 1 minute, unexpectedly showing apparently no degradation when the total RNA was analyzed using an Agilent BioAnalyzer. Using an alternative method to determine degradation, these were seen to contain degraded transcripts (see below).

Figure 3.Electropherogram of total RNA (~150 ng) resolved by capillary electrophoresis using the Agilent 2100 BioAnalyzer. The RNA had been incubated on a naked human hand for 1 minute prior to analysis. Analysis RIN of 10 indicated that the RNA was apparently high quality and intact.
For this reason it is important to apply caution to interpretation of RIN and other automatically generated measures of sample integrity and consider an additional, transcript-specific investigation. One such approach relies on measurement of a target using two independent assays. The integrity assay relies on measuring the ratio of the quantities of the target located towards the 5’ and at the 3’ of the same transcript (Figure 4)10. This provides a relative measure of transcript-specific RNA integrity (for a 3’/5’ assay protocol, see Appendix A).

Figure 4.Diagram to represent the 3’/5’ integrity assay. cDNA is produced from total RNA using oligodT primed RT. Two qPCR assays are designed to target the same transcript and the probes are labeled with different fluorophores, to facilitate multiplex detection. If the RNA is intact, detection of 5‘ and 3’ assays should be equal, whereas if the RNA is degraded there would be a higher detection of the 3’ assay relative to the 5’ assay.
In this way, the 3’/5’ ratio assay is used to evaluate the quality of RNA samples. The use of this approach is demonstrated in Figure 5, by testing the state of the GAPDH transcript (the specific targets under examination should be tested in any given experiment) in the RNA sample that was shown in Figure 3. After an oligo-dT primed cDNA synthesis, the GAPDH transcript is quantified using both a 5’ and 3’ assay. If the RNA is intact an equal concentration is expected using each assay (ratio = 1), whereas if the RNA is degraded a higher copy number is expected for the 3’ assay relative to the 5’ assay.
Figure 5. Demonstration of 3’/5’ Assay for Identification of Degraded RNA Samples. Although the Agilent 2100 BioAnalyzer analysis returned a RIN of 10 for both of these RNA samples, the 3’/5’ assay was used to demonstrate a 9 Cq (approximately 1,000-fold) loss in the 5’ assay for the sample after incubation for 1 minute on the human hand (1’ degradation) while the 3’ remained constant.
Analysis of the RNA using the Agilent 2100 BioAnalyzer led to the conclusion that the RNA shown in Figure 3 was intact, recording a RIN of 10 for both the RNA after 1’ incubation on the naked human hand and the same purified sample prior to degradation. When applying the 3’/5’ assay, the 3’ assay showed the same Cq for both samples, whereas the 5’ assay shows a loss of 9 cycles, indicating 1,000-fold loss of GAPDH 5’ and that the transcript is degraded. This observation supports the demonstration that the status of the rRNA (a significant component of the RIN algorithm) is not a reliable indicator of the integrity of the mRNA. However, the reliability of the 3’/5’ assay as a measure of overall sample quality depends upon the rate at which individual transcripts degrade11. To examine this, RNA extracted from HT29 cells was subjected to degradation by incubation for 72 hr at room temperature, or alternatively for 2 hr at 62 °C. All sample quality was analyzed using the Agilent 2100 BioAnalyzer. Assays were also designed to the 3’ and 5’ regions of 13 ubiquitously expressed genes and the quantity of these targets was determined in the purified, high quality undegraded RNA sample (HT29) using both assays for each gene (Figure 6A).

Figure 6A.RNA Quality Determination using 3’/5’ Assay a) RNA samples from HT29 cells with RIN approximately 10 were reverse transcribed using oligo-dT and the 3’/5’ ratio (y axis) determined for each of 13 genes (X axis). There is a general 3’ bias in quantification when both assays are used for the same target and the RNA is high quality. The degree of bias differs between genes, thus influencing the determination of ratios between genes, such as when normalizing to a reference gene. (Data kindly provided by Prof. Stephen Bustin, UK.)

Figure 6B.RNA samples from HT29 cells with RIN approximately 7 were reverse transcribed using oligo-dT and the 3’/5’ ratio (y axis) determined for each of 13 genes (X axis). There is a strong 3’ bias in quantification when both assays are used for the same target and the RNA is high quality and a clear 3’ shift from the ratio demonstrated in Figure 7.6A. The degree of the shift differs significantly between genes, indicating that different genes degrade at different rates. (Data kindly provided by Prof. Stephen Bustin, UK.)

Figure 6C.RNA samples from HT29 cells with RIN approximately 5 were reverse transcribed using oligo-dT and the 3’/5’ ratio (y axis) determined for each of 13 genes (X axis). There is a wide spread of data for replicates. (Data kindly provided by Prof. Stephen Bustin, UK.)
When using high quality RNA, the data for replicates are very close, but it is clear that for many genes there is a bias in the assay of up to 10 fold, usually towards the 3’ with a more dramatic 100-fold bias seen for UBC. This may reflect degradation or differential detection by the assays due to template folding or other RT-PCR influencing factor. Some targets appear to have a 5’ bias in detection which may be due to assay efficiency differences or other components of the RT-qPCR assay. The assays were then used to quantify the same targets using cDNA from RNA with RIN of 7.3 and 5.3 (Figures 6B and 6C, respectively). There were clear signs of degradation when using RNA samples of moderate degradation (RIN 7), with most genes showing a 3’ shift of between 10 and 1,000-fold indicating degradation of these transcripts between the 3’ and 5’ assays. Interestingly, three genes showed a 5’ shift which may indicate that degradation has resulted in a conformation change making the RT or 5’ qPCR more efficient. Since the changes for each gene are different it is clear that a ratio between genes measured using RIN 9 RNA would differ significantly from that measured using RIN 7 RNA. When using RNA of RIN 5 there is a higher degree of variability and the 3’/5’ ratio for each gene averages around 1. Since an average of 3’/5’ =1 would also be expected from high quality intact RNA, it is recognized that this system is applicable for integrity determination of RNA samples of moderate to apparently no degradation and is an improvement when used in addition to gel or capillary electrophoresis.
The observation of 5’ bias in the assays was intriguing. In theory, for assays of identical efficiency, it should not be possible to generate more 5’ than 3’ amplicon. Clearly additional factors are affecting the differential performance of these assays. A further sample quality consideration is the presence of contaminants, which may inhibit or even enhance RT or qPCR performance and whether these may be genetarget specific.
Contamination
Within the context of PCR assays, the significant forms of contamination are nucleic acid template that is inappropriately present in the sample, which can also be detected along with the specific target and result in a false positive, or material which can inhibit downstream reactions, causing false negatives or lower estimates of template concentration.
Template Contamination
PCR assays are especially vulnerable to amplicon contamination with the specific target sequence of interest because the process of PCR generates copies of the amplicon exponentially. Consequently, the process of performing a PCR assay produces the perfect contaminant for future PCR assays. In a molecular biology laboratory, it is essential to separate the space in which the reaction is set up and run from the post-PCR analysis area where the PCR product will be analyzed and further manipulated. It is preferable, where possible, to use separate rooms with dedicated equipment and laboratory coats such that nothing from the post-PCR analysis space is ever in contact with the clean, (pre-PCR) space. Many laboratories also introduce further controls for personnel access to these areas to prevent reaction carry over. Quantitative PCR assays are particularly vulnerable to amplicon contamination because they are capable of detecting a single template molecule and contaminating template will confound measurements of quantity.
In addition to PCR generation of the specific amplicon, care is needed in the laboratory setup to ensure that original template material is not transferred from one sample to another (cross-contamination) or, in the case of analysis of human samples, that material is not introduced from the personnel performing the analysis.
Adopting a standard experimental procedure with appropriate controls to identify potential sources of contamination is highly recommended.
Contamination of RNA with gDNA
When analyzing RNA, it is important to ensure that there is no gDNA contamination. Enzymatic treatment of the samples with DNase I in solution is recommended for removing contaminating gDNA and is particularly important for investigations of intron-less genes or when the assay design does not distinguish between the gDNA and cDNA sequences. Wherever possible, it is advisable to design assays over the intron/exon boundary such that only the processed mRNA sequences can be amplified and/or detected. This does not always prevent amplification from gDNA, as there are sequences that exist as processed pseudo genes which are effectively genomic cDNA sequences (see PCR/qPCR/dPCR Assay Design). It is also worth considering that DNA exists in the reagents and potentially, can cause contamination issues12.
Inhibition
The presence of inhibitors differentially affects qPCR assays to different targets13. Therefore, the presence of inhibitors in a sample can result in complex perturbations of the true data. Since each assay is affected differently, the resulting ratios between gene of interest (GOI) and reference gene quantities may not reflect the true abundance of each gene, leading to incorrect estimates of relative target quantities or difficulties in interpreting genotyping assays.
PCR inhibitors that are commonly introduced during experimental processes include; tris, ethanol, isopropanol, EDTA, the reverse transcriptase enzyme, guanidine isothiocyanate and phenol.
One system that can be used to detect inhibitors is the SPUD assay14 (Figure 7). Using this method, an artificial amplicon (derived from a potato nucleic acid sequence lacking homology with any other known sequence) is subjected to qPCR in the presence of either water (amplification control; Figure 7 sample a) or the test sample (Figure 7 samples b and c). If the test sample does not contain an inhibitor of the SPUD assay, the quantification cycle (Cq) recorded for the amplification control and test sample will be the same. However, if the test sample contains an inhibitor, the Cq will be increased (a detailed protocol for the test is provided as the SPUD Protocol, Appendix A).

Figure 7.7.Representation of the SPUD assay. In this case, the difference in Cq between the sample c and the control sample a shows presence of an inhibitor.
An Integrated Example: Analysis of RNA Sample Quality
Each of the sample quality control methods described previously is used to determine a specific feature of the sample. This section contains a description of an example workflow that may be applied to samples, with a description of the data that each quality control test provides. The presented investigation is neither exhaustive nor provided as a definitive standard operating procedure; it is merely provided as an example of a possible quality control procedure.
Five RNA samples have been prepared and are labeled sequentially A to E. These were analyzed using the NanoDrop and were found to be of very similar concentration (around 100 ng/μL). The A260/A280 ratio was recorded twice using two independent readings for each sample (Figure 8) and was found to be approximately 1.9 in all cases, suggesting samples of high quality and without contaminants as would be detected at A280 nm, it is noteworthy that an A230 nm reading is absent from this analysis. An analysis using capillary electrophoresis (the Agilent 2100 BioAnalyzer) reveals a different picture (Figure 9). Samples A and B are of high quality (RIN 10 and concentration 110 ng/μL in agreement with NanoDrop reading), sample C is highly degraded (RIN 2.4, concentration 62 ng/μL is half the concentration determination of the NanoDrop) and samples D and E are high quality (RIN 9.1 and 9.5, respectively) but of lower concentration than samples A and B (30 ng/μL and 43 ng/μL, respectively). The information from the two methods of measurement is somewhat complementary because the NanoDrop does not reveal the degradation. However, it is also in conflict because the concentration of the samples D and E differ significantly. This would be cause for concern and should trigger further investigations. While capillary electrophoresis is a powerful tool for sample evaluation, it is relatively low throughput and not available to all researchers. An alternative test for sample integrity is the 3’/5’ analysis. This test was applied to samples A–E using equal concentrations according to the NanoDrop reading. When these samples were tested using this approach, it was clear that the RNA of samples A (Figure 10A) and B (data not shown, identical to sample A) were intact, whereas that for sample C was highly degraded (Figure 10B). Note that for sample C the Cq for both the 5’ and the 3’ assays has shifted significantly to higher values demonstrating a loss of specific target relative to samples A and B. The profile for samples D and E was not in accordance with expectations (Figure 10C shows sample D, data is not shown for sample E but was similar) since the 5’ assay has a lower Cq than the 3’ indicating more 5’ signal than 3’ in a cDNA sample produced from oligo-dT priming. The alternative explanation is that these samples contain an inhibitor which affects the 3’ assay more than the 5’. To test this theory, the SPUD assay was run on all samples (Figure 11). There is no evidence of an inhibitor in samples A to C but both samples D and E show inhibition of the SPUD assay, with sample D causing the assay to fail completely.
Using all of the information, it can be concluded that there was an equal concentration of nucleic acid material in each sample (NanoDrop) and the samples contained undetectable contaminants that absorbs at 280 nm (protein) (other wavelengths were not tested). Samples A and B were of high integrity without detected inhibitors, sample C was highly degraded (capillary electrophoresis and 3’/5’ assay) without detected inhibitors and samples D and E were intact but contained PCR inhibitors (3’/5’ and SPUD).

Figure 8.NanoDrop Quality Assessment (A260/A280).

Figure 9.Agilent 2100 BioAnalyzer Analysis of RNA Samples A–E.
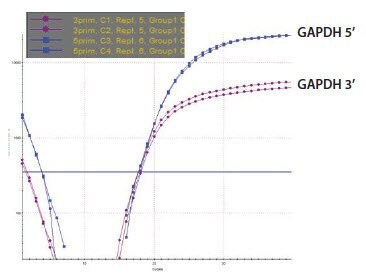
Figure 10A.GAPDH 3’/5’ Multiplex Assay RNA Sample.

Figure 10B.GAPDH 3’/5’ Multiplex Assay Sample C.

Figure 10C.GAPDH 3’/5’ Multiplex Assay Sample D.
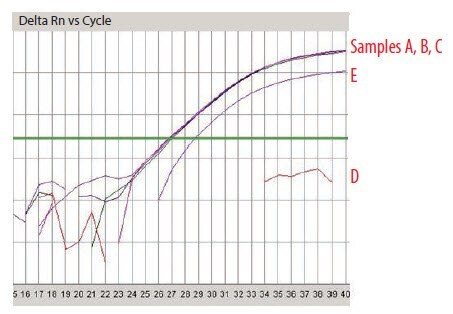
Figure 11.SPUD Analysis of Samples A–E.
References
Per continuare a leggere, autenticati o crea un account.
Non hai un Account?