Practical Considerations for IEX Separation
Extracted from Ion Exchange Chromatography & Chromatofocusing, GE Healthcare, 2007
This section covers detailed aspects of each step in an IEX separation, together with practical hints and tips to improve resolution and overall performance. In practice a separation can be summarized as follows:
- Equilibrate column with 5–10 column volumes of start buffer or until the baseline, eluent pH and conductivity are stable.
- Adjust the sample to the chosen starting pH and ionic strength and apply to the column.
- Wash with 5–10 column volumes of start buffer or until the baseline, eluent pH and conductivity are stable i.e. when all unbound material has washed through the column.
- Begin elution using a gradient volume of 10–20 column volumes with an increasing ionic strength up to 0.5 M NaCl (50%B). Alternatively (if gradient-making equipment is not available) elute bound proteins with 5 column volumes of start buffer + NaCl at chosen ionic strength. Repeat at higher ionic strengths until the target protein(s) has been eluted.
- Wash with 5 column volumes of 1 M NaCl (100%B) to elute any remaining ionically bound material.
- Re-equilibrate with 5–10 column volumes of start buffer or until eluent pH and conductivity reach the required values.
These steps are highlighted throughout this section.
Buffer volumes referred to are expressed in column volumes, for example 3 CV=3 mL for a column with a 1 mL bed volume. Using column volumes to describe a separation profile facilitates method development and transfer of methods to columns of different dimensions.
The number of column volumes used at each stage of the separation can often be reduced by optimization. For example, less buffer is required to equilibrate a strong ion exchanger, the gradient volume can be reduced if resolution can be maintained and less buffer may be required for washing when separating less complex and reasonably clean samples.
pH and Ionic Strength
Buffer pH and ionic strength must be compatible with protein stability and activity. The most suitable pH should allow the proteins of interest to bind, but should be as close to the point of release (elution) as possible. If the pH is too low or too high, elution becomes more difficult and high salt concentrations may be needed. This should be avoided since some proteins begin to precipitate at high ionic strength and high salt concentrations may interfere with assays or subsequent chromatographic steps.
Avoid extreme changes in pH or other conditions that may cause inactivation or even precipitation.
The pH and ionic strength of the sample are extremely important in order to achieve the most effective high resolution or group separations and to make the most of the high loading capacity. Ideally, samples should be in the same conditions as the start buffer (Appendix 1, Sample preparation and, in particular, Buffer exchange and desalting, page 156 for details). When working with small volumes during screening and scouting, it may be sufficient to dilute the sample in start buffer in order to lower the ionic strength and adjust the pH to a value similar to that of the start buffer.
Proteins often begin to dissociate from IEX media about 0.5 pH units from their isoelectric points at an ionic strength around 0.1 M. The pH of the start buffer should be at least 0.5–1 pH unit above the pI of the target substance when using an anion exchanger (Q, DEAE or ANX) or 0.5–1 pH unit below the pI of the target substance when using a cation exchanger (SP or CM).
For samples with unknown charge properties, try the following:
- anion exchange (Q, DEAE or ANX)
start buffer: pH 8.0
elution buffer: start buffer including 1 M NaCl, pH 8.0 - cation exchange (S, SP, CM)
start buffer: pH 6.0
elution buffer: start buffer including 1 M NaCl, pH 6.0
Appendix 2 for recommendations on volatile and non-volatile buffer systems for anion and cation exchangers.
Whenever possible, check for stability at the pH and ionic strength values selected, especially if recovery of biological activity is a priority.
Anion or Cation Exchanger
For molecules such as nucleic acids which carry only negatively-charged groups, an anion exchanger is the obvious choice. However, since the net charge of molecules such as proteins (carrying positively and negatively charged groups) depends on pH, the choice is based on which type of exchanger and pH give the desired resolution within the constraints of sample stability. For example, Figure 18 shows a theoretical protein which has a net positive charge below its isoelectric point and can bind to a cation exchanger. Above its isoelectric point the protein has a net negative charge and can bind to an anion exchanger. However, the protein is only stable in the range pH 5–8 and so an anion exchanger has to be used.

Figure 18.Considerations when selecting a suitable IEX medium
If sample components are most stable below their isoelectric points, use a cation exchanger.
If sample components are most stable above their isoelectric points, use an anion exchanger.
If stability is high over a wide pH range on both sides of the isoelectric point, use either type of ion exchanger.
Strong or Weak Ion Exchangers
Table 3 shows the functional groups used on IEX media. The terms strong and weak refer to the extent that the ionization state of the functional groups varies with pH. The terms strong and weak do not refer to the strength with which the functional groups bind to proteins.
Begin with a strong exchanger to enable development work to be performed over a broad pH range. Use a strong anion exchanger (Q) to bind the protein(s) of interest if their isoelectric point is below pH 7.0 or unknown.
Use a strong exchanger in those cases where maximum resolution occurs at an extreme pH and the proteins of interest are stable at that pH.
Consider using a weak exchanger if the selectivity of the strong ion exchanger is unsatisfactory, but remember that the ion exchange capacity of a weak ion exchanger varies with pH. As a result:
- sample loading (binding) capacity can vary with increasing pH due to loss of charge from the exchanger.
- resolution is more readily affected by changes in flow rate or sample load due to the intermediate forms of charge interaction which can occur.
- predicted results (based on known information about the sample components such as their isoelectric points and how their net surface charge changes with pH) may not correlate with actual results since the number of charged groups on weak ion exchangers can vary with pH.
- longer equilibration times may be required in order to titrate the weak ion exchange functional groups.
When using a weak exchanger, work within the pH values given below to minimize variations in performance:
DEAE: pH 2–9
ANX: pH 2–9
CM: pH 6–10
Buffer Selection and Preparation
Buffer ions
Buffering ions should have the same charge as the functional groups on the IEX medium (buffering ions that carry a charge opposite to that of the functional groups will take part in the ion exchange process and can cause significant pH fluctuations during elution) and, preferably, a pKa value within 0.6 pH units of the working pH. An exception to this rule is seen in the frequent use of phosphate buffers with anion exchange separations. However, phosphate buffers must be very carefully prepared to ensure reproducibility between batches.
Use a buffer concentration that is sufficient to maintain buffering capacity and constant pH, typically 20–50 mM.
Use volatile buffers if the purified product is to be lyophilized.
Appendix 2 for recommendations on volatile and non-volatile buffer systems for anion and cation exchangers.
Filter buffers after all salts and additives have been included. Use high quality water and chemicals. Filter solutions through 1 μM filters for particle sizes above 90 μM, 0.45 μM filters for 34 μM particles or 0.22 μM filters for particles sizes below 15 μM or when sterile or extra clean samples are required. To avoid formation of air bubbles in a packed column, ensure that column and buffers are at the same temperature when preparing for a run.
Effect of temperature on buffer pH
Select buffers that have appropriate pKa values for the working temperature. The pKa of a buffering substance varies with temperature. For example Tris™ has a pKa of 8.85 at 0 °C, 8.06 at 25 °C and 7.72 at 27 °C. Using Tris at 4 °C at a pH 7.9 would give a very low buffering capacity and the working pH would be outside the useful pH range (pKa + 0.5) of the buffer.
Prepare buffers at the same temperature at which they will be used.
Temperatures <10 °C can minimize aggregation caused by hydrophobic interactions between sample components. Working at these lower temperatures may be an alternative solution to using a detergent to improve solubility.
Counter-ions
The counter-ions (salt ions) used in IEX are almost always Na+ for cation exchange and Cl– for anion exchange.
Salts such as NaCl have a chaotropic character (i.e. an ability to make water less polar) and therefore a lower 'salting-out' effect on hydrophobic molecules. This ensures maximum solubility during elution and improves recovery. Chaotropic salts can also be used in the presence of organic solvents if required. Salts such as (NH4)2SO4 or K3PO4 should be avoided as they are most likely to cause precipitation at high concentrations.
In certain applications alternative counter-ions such as Li+, Br–, I–, SO42–, CH3COO– or
HCOO– may improve and even alter, selectivity since they exhibit different elution strengths, but it should be noted that using these ions may affect the binding capacity of the medium.
Figure 19 shows how selectivity and resolution can vary when using different counter-ions.

Figure 19.Effect of counter-ions on selectivity and resolution (Mono Q HR 5/5 now available as Mono Q 5/50 GL). Note the variation in elution order of peaks 3 and 4.
Use the following procedure if a medium is to be used with counter-ions other than sodium or chloride:
- Wash the packed column with 10 column volumes 0.5–1 M salt solution containing the new counter-ion. Flow rate: see relevant media section in Chapter 3.
- Wash with 10 column volumes of start buffer at the same flow rate as in step 1.
- Repeat steps 1 and 2 several times.
Perform a blank run to check conductivity and pH.
Column and Media Preparation
Equilibrate column with 5–10 column volumes of start buffer or until the baseline, eluent pH and conductivity are stable.
Using prepacked columns is highly recommended to ensure the best performance and reproducible results. An evenly packed column ensures that component peaks are not unnecessarily broadened as sample passes down the column so that the best resolution can be achieved.
Allow buffers, media or prepacked columns to reach the same temperature before use. Rapid changes in temperature, for example removing packed columns from a cold room and then applying buffer at room temperature, can cause air bubbles in the packing and affect the separation.
Wash away storage solutions and preservatives before using any IEX medium.
Increase the volumes used for column equilibration before the first run if using buffers containing detergents or a different counter-ion to the one in which the medium has been stored.
Appendix 3 gives details on column packing. The volume required for the packed bed is determined by the amount of sample to be purified and the binding capacity of the medium. Pack a column that will have approximately 5-fold excess of the binding capacity required with a bed height up to 20 cm.
Check column performance regularly by determining column efficiency and peak symmetry. Appendix 3. Note that this does not apply to HiTrap or HiPrep™ columns.
Sample Preparation
Correct sample and buffer preparation is essential in order to achieve optimal separation and avoid any deterioration in column performance. Simple steps to clarify a sample before application to a column will avoid the risk of blockage and reduce the need for stringent washing procedures. Appendix 1 contains a detailed overview of sample preparation techniques.
Desalt samples and transfer into the chosen start buffer (refer to page 156 for details of buffer exchange and desalting). The pH and ionic strength of the sample are extremely important in order to achieve the most effective high resolution or group separations and to make the most of the high loading capacity.
For small sample volumes in a high salt concentration and with no major contaminants such as lipids or ionic detergents, it may be sufficient to dilute the sample with start buffer in order to lower the salt concentration to a level that does not interfere with binding to the medium. However, buffer exchange and desalting is the only way to guarantee the correct pH and ionic strength conditions of a sample.
Samples must be clear and free from particulate matter, particularly when working with particle sizes of 34 μM or less. For small sample volumes, a syringe-tip filter of cellulose acetate or PVDF can be sufficient for sample filtration.
Concentration and viscosity
The solubility or viscosity of the sample may limit the quantity that can be applied to a column. High sample viscosity can cause instability of the separation and an irregular flow pattern resulting in broad, distorted peaks and problems with back pressure. The critical parameter is the viscosity of the sample relative to the viscosity of the eluent.
Dilute viscous samples with start buffer. If high viscosity is caused by the presence of nucleic acid contaminants, Appendix 1 for advice on their removal. Remember that viscosity varies with temperature. If dilution is not an option, using a medium with a larger particle size may help to overcome viscosity problems.
Samples should generally not exceed 50–70 mg/mL protein, but may vary according to the type of sample and the type of chromatographic medium.
Sample Application
Adjust the sample to the chosen starting pH and ionic strength (sample preparation) and apply to the column.
Wash with 5–10 column volumes of start buffer or until the baseline, eluent pH and conductivity are stable i.e. when all unbound material has washed through the column.
Starting conditions should maximize binding of the target proteins near the top of the column and, when possible, minimize binding of contaminants so that they pass through the column.
For efficient binding the sample should be at the same pH and ionic strength as the start buffer. The sample volume can be relatively large without affecting the separation since the sample will bind at the top of the column as long as equilibration and sample conditions are correct.
Apply samples directly to the column via a chromatography system, a peristaltic pump or a syringe. The choice of equipment depends largely on the sample volume, the size of column, the type of IEX medium and the requirements for accuracy in gradient elution. Ensure that the top of the column bed is not disturbed during sample application
Do not change buffer conditions until all unbound material has been washed through the column (monitored by UV absorbance) and until UV and conductivity values have returned to starting conditions.
Sample Load
Sample load (mass) is of greater importance than sample volume. The amount of sample which can be applied to a column depends on the dynamic binding capacity of the IEX medium and the degree of resolution required. Sample load has a major influence on resolution since the width of the peaks is directly related to the amount of substance present, as shown in Figure 20. Consequently, in order to achieve satisfactory resolution, the total amount of protein applied and bound to the medium should not exceed the total binding capacity of the packed column.

Figure 20.The influence of increasing sample load on resolution.
Apply up to 30% of the total binding capacity of the column for optimal resolution with gradient elution. Sample loads can be increased if resolution is satisfactory or when using a step elution.
If sample volumes are large compared to the total column volume, the sample buffer composition, in particular the ionic strength, should be the same as that of the start buffer to ensure adequate binding conditions.
Chapter 3 gives typical binding capacities for each medium as a guideline for total binding capacity. The actual (dynamic) binding capacity is also affected by factors such as size and shape of the molecules, the pore size of the matrix, flow rate, sample concentration, pH/protein charge and ionic strength. Capacity will decrease for molecules of very large diameter or length, e.g. protein complexes >Mr 400 000, assymmetric proteins and DNA. These molecules are unable to penetrate the matrix pores, limiting their binding primarily to the charged groups on the surface of the matrix. Since the exact distribution of pore sizes in some matrices can vary and the apparent size of a molecule can vary according to the buffer conditions, there is no distinct molecular weight cut-off point when molecules can or cannot penetrate the matrix pores.
The binding step and the dynamic binding capacity can be increased by applying sample at a pH where the target protein has a higher charge than if the optimal pH for separation was used.
Sample volume
As a binding technique, IEX is independent of sample volume as long as the ionic strength of the sample is the same or as low as the start buffer and the target proteins are sufficiently charged at the selected pH. Large volumes of dilute solutions, such as fractions from a desalting step or a cell culture supernatant, can be applied directly to an IEX medium without prior concentration.
Elution
Bound proteins are eluted by controlled changes in ionic strength or pH. The way in which these changes take place, by using a linear or step elution, is selected according to the aim of the separation:
- Linear gradient elution
– high resolution separation or analysis
– optimized gradient elution at increased speed while retaining required resolution - Step elution
– faster separation time, reduced buffer consumption
– group separation
Linear Gradient Elution
Aim: high resolution separation or analysis, screening
Begin elution using a linear gradient volume of 10–20 column volumes with an increasing ionic strength up to 0.5 M NaCl (50%B).

Figure 21.Typical IEX separation using linear gradient elution. The UV (protein) and conductivity (salt) traces show the elution of protein peaks and the changes in salt concentration during elution.
Linear ionic strength gradients, as shown in Figure 21, are the most frequently used type of elution and should always be used when starting with an unknown sample (when as many components as possible are bound to the column and eluted differentially to see a total protein profile). At low ionic strengths, competition for charged groups on the IEX medium is at a minimum. Increasing the ionic strength increases competition and reduces the interaction between the medium and the bound substances which begin to elute. The elution buffer is usually the same buffer salt and pH as the start buffer, but contains additional salt, most often sodium chloride.
It is strongly recommended to use linear gradient elution during method development. Linear ionic strength gradients are easy to prepare and very reproducible when generated by a suitable chromatography system. The results obtained can then serve as a base from which to optimize the separation.
The retention of charged proteins on the medium is related to the volume of the column and the concentration difference across it:
- long, shallow gradients give maximum separation between peaks, but separation times will be longer and there will be greater peak broadening.
- short, steep gradients give faster separations and sharper peaks, but peaks will be eluted closer together.
- peaks eluted later in the gradient tend to be slightly broader than those eluted early on.
Select the steepest gradient to give acceptable resolution at the selected pH.
The effects of gradient slope are shown in Figure 22.

Figure 22.Effect of gradient slope on resolution, in theory and in practice.
If gradient elution volumes are decreased, it may be necessary to decrease the sample load proportionally in order to maintain the same resolution. Similarly, if sample load is increased (within the total capacity of the column), gradient volumes may need to be increased to maintain resolution.
Gradients are best formed using purpose-designed equipment, such as ÄKTAdesign systems with preprogrammed method templates, that automatically controls the mixing of solutions being supplied to a column. Users of ÄKTAdesign systems with BufferPrep functionality can select from a range of buffer recipes to run salt gradient elutions at constant pH.
BufferPrep automatically calculates and mixes the correct proportions of stock solutions in order to maintain a constant pH throughout the run. Alternatively, systems may use two separate pumps for start and elution buffers or a single pump in combination with a switch valve to mix the buffers.
Accurate buffer preparation, efficient mixing and the shortest possible flow path between a mixer and the top of a column will help to ensure accurate gradient formation.
Aim: reduced separation time, maintained resolution
For certain separations, when conditions for a high resolution separation using a linear gradient have been established, it may be possible to reduce the total separation time by using a more complex elution profile, as illustrated in Figure 23. Shallow gradients can be used where maximum resolution is required while steeper gradients can be used in areas where resolution is satisfactory.
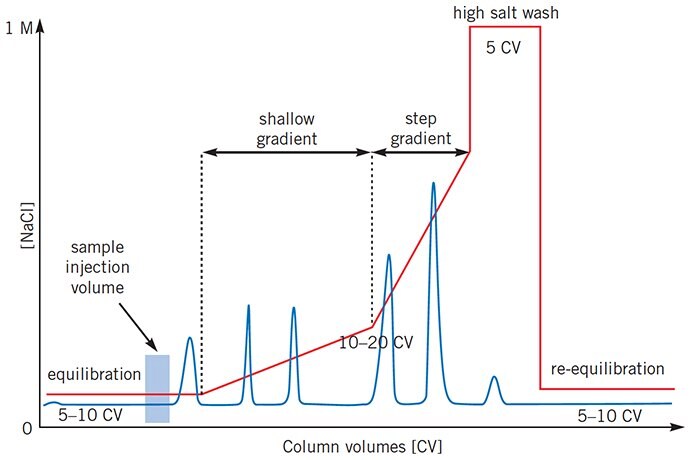
Figure 23.Complex gradient profiles can reduce total separation time for certain separations.
Step Elution
Elute bound proteins with 5 column volumes of start buffer + NaCl at chosen ionic strength.
Repeat at higher ionic strengths until the target protein(s) has been eluted.

Figure 24.Typical IEX separation using step elution. The UV (protein) and conductivity (salt) traces show the elution of protein peaks and the changes in salt concentration during elution.
As shown in Figure 24, step elutions are performed by sequential addition of the same buffer at increasing ionic strengths. Step elution is technically simple, but care must be taken in the design of the steps and the interpretation of results since substances eluted by a sharp change in ionic strength elute close together, giving a false peak that may contain several components. Peaks tend to have sharp fronts and pronounced tailing since they frequently contain more than one component. Tailing may lead to the appearance of false peaks if a change in ionic strength is introduced too early. For these reasons it is recommended to use a linear ionic strength gradient when developing a new method.
Aim: faster separation time, reduced buffer consumption
When an IEX separation has been optimized using gradient elution, changing to a step elution reduces the total number of column volumes used for a separation. This speeds up separation times and reduces buffer consumption while retaining the required purity level. Step elutions of this type are often used for routine, large scale separation. An added advantage of a step elution when used at larger scale is that it is often possible to apply a greater amount of sample, since the molecules which would elute early in a gradient separation no longer take up binding capacity on the column.
Aim: group separation
In a group separation the molecules of interest are concentrated and rapidly removed from unwanted substances. When binding and elution conditions for a target protein(s) and contaminants have been determined, usually during preliminary gradient elution separations, conditions are chosen to maximize binding of the target protein(s) and minimize binding of contaminants during sample application. The target protein(s) is then eluted by a single buffer change in an enriched, concentrated form. Figure 25 shows an example of such a separation in which a HiTrap Q HP column is used to separate human serum proteins from the unwanted IgG fraction which passes directly through the column.
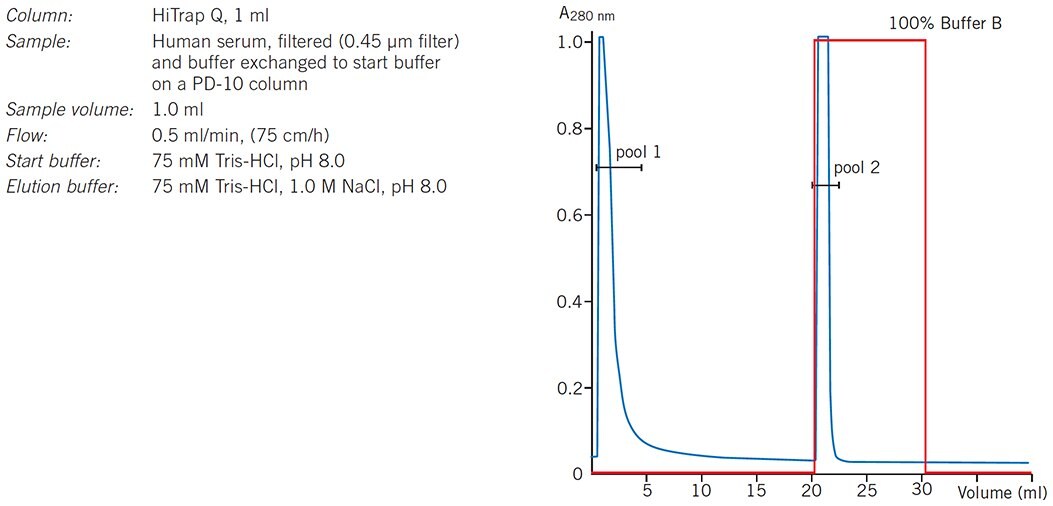
Figure 25.Group separation of serum proteins on HiTrap Q HP.
Aim: removal of contaminants
If starting conditions have been chosen to maximize the binding of contaminants then no change in elution conditions is required since the target protein(s) will pass through the column. For many applications it is preferable to discard the column rather than spend time and effort removing unwanted bound substances.
pH elution
Since the net charge on a protein is pH dependent, samples can also be eluted from an IEX medium by altering the pH of the elution buffer. As there is no salt gradient, samples are simply retained on the column at one pH and eluted by increasing or decreasing the pH. The various charged groups in the sample or on the column are titrated until they are neutral or of opposite charge to the medium and the sample elutes.
- Proteins bound to an anion exchanger (Q, DEAE, ANX) will elute as pH is decreased.
- Proteins bound to a cation exchanger (SP, S, CM) will elute as pH is increased.
Since pH elution will involve working at pH values close to the isoelectric point of a protein and since many proteins show minimum solubility close to their isoelectric points, precautions must be taken to avoid precipitation on the column (page 48 for information on the use of additives to avoid precipitation).
Always test in advance the solubility of sample components at the pH and salt concentrations to be used during separation.
For any type of pH elution, care must be taken in the selection and mixing of buffer systems in order to achieve reproducibility. Stepwise pH elution is easier to produce and more reproducible than using a linear pH gradient. Note that for weak ion exchangers the buffer may have to titrate the charged groups on the medium and there will be a short period of re-equilibration before the new pH is reached.
Linear pH gradients are very difficult to produce at constant ionic strength, since simultaneous changes in ionic strength, although small, also occur. These gradients cannot be obtained simply by mixing buffers of different pH in linear volume ratios since the buffering capacities of the systems produced are pH dependent. A relatively linear gradient can be produced over a narrow pH interval (maximum 2 pH units) by mixing two solutions of the same buffer salt adjusted to 1 pH unit above and 1 pH unit below the pKa for the buffer.
In general, separation of proteins according to their isoelectric points, using chromatofocusing (Chapter 5), is likely to provide a more reliable and higher resolution result than attempting to elute proteins from an IEX column using a pH gradient.
Flow Rates
The maximum flow rate applied during a separation can vary according to the stage of the separation. For example, during sample application and elution, lower flow rates allow time for sample components to diffuse in and out of the pores as they to bind to or dissociate from the functional groups. Figure 26 shows an example of the influence of flow rate on resolution. Higher flow rates can be used for equilibration, washing and re-equilibration, limited primarily by the rigidity of the media and by pressure specifications of the equipment.
Recommended flow rates for each chromatography medium are given in Chapter 3. Working from these recommendations, select the highest flow rate that maintains resolution and minimizes separation time. For example, if peaks are well separated at a low flow rate, increase the flow rate or, alternatively, increase the sample volume to benefit from a higher capacity without significant loss of resolution.

Figure 26.Influence of increasing flow rate on resolution.
Flow rate is measured in simple volume terms, e.g. mL/min, but when comparing results between columns of different sizes or when scaling-up, it is useful to use linear flow: cm/hour (Appendix 5). Results obtained at the same linear flow on different size columns will be comparable as far as the effects of flow rate are concerned.
Save time by using higher flow rates during the high salt wash and re-equilibration steps. Do not exceed the maximum recommended flow for the medium.
Higher flow rates and viscous buffers increase operating pressures (remember that buffer viscosity increases when running at +4 °C). Check the maximum operating pressure of the packed column and set the upper pressure limit on the chromatography system accordingly.
Flow Control
Accurate, reproducible flow control is essential for good resolution and reproducibility.
Use a pump within a chromatography system (rather than a peristaltic pump) to fully utilize the high rigidity and excellent flow properties of media such as MiniBeads, MonoBeads, SOURCE or Sepharose High Performance.
Always pump the buffer onto a column (rather than drawing the buffer through the column with the pump below). This reduces the risk of bubble formation as a result of suction. If you have packed the column yourself, always use a flow rate for separation that is less than the flow rate used for column packing in order to avoid shrinking of the column bed by pressure increases that may occur when running a sample.
Wash and Re-equilibration
Wash with 5 column volumes of 1 M NaCl (100%B) to elute any remaining ionically-bound material.
Include a wash step at the end of every run in order to remove any molecules that are still bound to the medium. Monitor UV absorbance so that the wash step can be shortened or prolonged, as necessary.
Re-equilibrate with 5–10 column volumes of start buffer or until eluent pH and conductivity reach the required values.
A re-equilibration step after washing returns the column to start conditions before applying further samples. Whenever possible, monitor pH and conductivity to check when start conditions have been reached. The re-equilibration step can then be shortened or prolonged as necessary.
Increase flow rates during wash and re-equilibration steps to save time between runs.
If ionic detergents have been used, wash the column with 5 column volumes of distilled water, followed by 2 column volumes 2 M NaCl. Re-equilibrate with at least 10 column volumes of start buffer until the UV baseline, eluent pH and/or conductivity are stable. Organic solvents such as ethanol can be used to remove non-ionic detergents. When selecting an organic solvent, check the chemical stability of the medium to determine a suitable concentration.
Detergents, Denaturing Agents, and Other Additives
Any additives used for dissociation, solubilization, metal chelation, enzyme inhibition etc. should always be checked for their charge characteristics at the working pH. Run blank gradients with additives included in order to check their effect on the chromatographic profile.
Additives used during sample preparation will be separated from the sample components during IEX. If proteins are seen to precipitate, elute later than expected or are poorly resolved, add a suitable concentration of the additives used for initial solubilization to the start and elution buffers.
Zwitterionic additives such as betaine can prevent precipitation and can be used at high concentrations without interfering with the gradient elution.
Detergents are useful as solubilizing agents for proteins with low aqueous solubility such as membrane components. Anionic, cationic, zwitterionic and non-ionic (neutral) detergents can be used during IEX chromatography.
Denaturing agents such as guanidine hydrochloride or urea can be used for initial solubilization of a sample and during separation. However, they should be avoided unless denaturation is a requirement. Note that, at the pH values used for separation, guanidine is a charged molecule with a counter-ion and will therefore participate in the ion exchange process in the same way as NaCl.
Examples of commonly used detergents and denaturing agents are given in Table 4.
Developing or optimizing a separation using buffers that contain detergents
- Select detergents that are compatible with the sample. A detergent must be neutral, zwitterionic or have the same charge as the IEX medium. Detergents that bind to the medium can be difficult to remove and may affect protein loading capacity, pH, conductivity and resolution.
- Determine the minimum concentration that is likely to keep the sample in solution during the separation. Note that different detergents will have different solubilization properties resulting in different peak profiles.
- Equilibrate the column thoroughly with the detergent solution, using a concentration that is below the critical micelle concentration for the specific detergent.
- Run blank salt gradients to determine the UV absorbance profile of the detergent and to detect any effect pH. Micelle formation causes light scattering and the appearance of a peak during UV monitoring. If micelle formation is a problem try the following:
- decrease detergent concentration as far as possible without impairing sample solubility
- increase detergent concentration to run the gradient above the critical micelle concentration (this creates a gradual rather than
abrupt UV increase)
- change the salt gradient so that the sudden change in UV absorption does not occur during the run
- change to highly chaotropic salts such as LiClO4 or sodium trichloroacetate that can be used at higher concentrations without
causing micelle formation - Perform test runs with sample to find the detergent that gives the best solubilization and resolution.
A single peak obtained from a 'detergent run' often contains more than one component and should be analyzed carefully. Selecting a different detergent may improve the separation.
Detergent concentrations that are too high will increase buffer viscosity so that flow rates must be reduced to avoid over-pressure of the column.The concentration of detergent required for solubilization can often be reduced during the separation.
Use detergents of the highest quality that are free from salts. Filter buffers that contain detergents under weak suction and ultrasonication for degassing in order to avoid foaming.
Wash previously used columns thoroughly using recommended procedures before working with buffers that contain detergents.
Reagents to reduce polarity
Monoethylene glycol, glycerol and similar mild reagents that reduce polarity can be included in buffers. Avoid high concentrations (>40% w/w) as buffer viscosity will increase and may over-pressure the column.
Metal chelators: EDTA, EGTA
EDTA (ethylenediaminetetracetic acid) and EGTA (ethylene glycol-bis-(2-aminoethyl)- N,N,N',N'-tetraacetic acid) are often used in buffers as metal chelators and can be used with IEX chromatography. EDTA and EGTA contain several carboxylic acid groups that may interact with anion exchangers. During anion exchange separations EDTA and EGTA can concentrate as a band on the column and elute during a salt gradient. Both molecules absorb UV and will appear as a peak or as background noise in the chromatogram.
To continue reading please sign in or create an account.
Don't Have An Account?