RNA Immunoprecipitation Chip (RIP) Assay
Protein-RNA interactions play important roles in the cell including structural, catalytic, and regulatory functions. Similar to chromatin immunoprecipitation (ChIP), RNA immunoprecipitation (RIP) can be used to detect the association of individual proteins with specific nucleic acids such as mRNAs, noncoding RNAs (e.g. long non-coding RNAs, enhancer RNAs, miRNAs), and viral RNAs. There are two major RIP assay variants: native and cross-linked. The native RIP assay allows for direct identification of bound RNAs to the protein of interest and their abundance in the immunoprecipitated sample. Whereas cross-linked RIP assays allow for the precise mapping of the direct and indirect binding site of the protein and RNA (Figure 1). For cross-linked RIP, live cells are treated with highly reactive formaldehyde to generate protein-RNA reversible cross-links between molecules that are in close proximity in vivo (Figure 2).
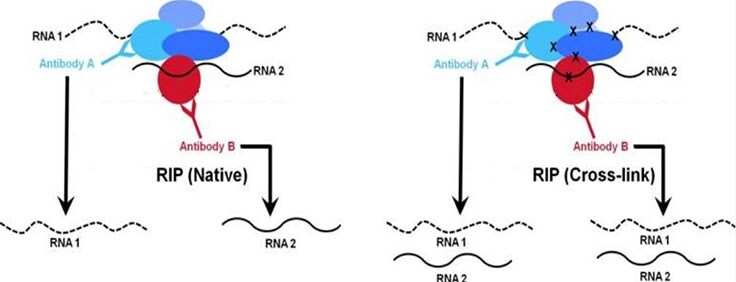
Figure 1. Comparison of native vs. cross-linkedWhile both native and cross-linked approaches are used to analyze chromatin-associated RNA, the native RIP approach (right panel) typically allows recovery of high affinity protein-RNA interactions. Conversely, the cross-linked method (right panel) is designed to capture higher molecular weight complexes and more readily trap weaker interacting RNAs.
https://www.emdmillipore.com/US/en/product/EZ-Magna-RIP-RNA-Binding-Protein-Immunoprecipitation-Kit,MM_NF-17-701RNA immunoprecipitation (RIP) Assay protocol workflow
The protocol that you select for you RIP assay will in part be determined by the protein-DNA binding location (cytoplasm or nuclear). A general whole cell lysate RIP protocol may be best suited for cytoplasmic interactions where a nuclear RIP protocol may be optimal for interactions occurring in the nucleus. If there is no existing knowledge about the location of the binding RNAs, the general whole cell lysate native RIP protocol (Figure 2A) (17-700, 17-701) is recommended to improve the possibility of immunoprecipitation of the binding RNAs regardless of their location. This protocol works with varied amounts and types of starting material, such as cells and tissue samples. If the binding RNAs are known to occur in the nucleus or are chromatin associated, the nuclear native (17-10522, 17-10523) or cross-linked RIP protocol (17-10520, 17-10521) is recommended. Instead of immunoprecipitating RNAs from the whole cell lysate, these protocols carry out the immunoprecipitation on the isolated nuclei, thus enabling improved signal-to-noise ratios.
While these protocols are designed for immunoprecipitation of all types of RNAs, there are two additional protocols specifically developed for identification and analysis of regions of chromatin that interact with specific chromatin-associated long non-coding RNAs/Chromatin Isolation by RNA Purification (ChIRP) (Figure 2B) (17-10494, 17-10495) or transcriptome-wide profiling of m6A (N6-methyladenosine) RNA methylation sites (Figure 2C) (17-10499).

Figure 2. Workflows of different RIP protocolsA. Whole cell lysate native RIP workflow (Product Nos. 17-700, 17-701) B. ChIRP workflow (Product Nos. 17-10494, 17-10495) C. m6A RIP workflow (Product Nos. 17-10499)
RIP (Assay protocol workflow) downstream analyses
RNAs isolated upon completion of a RIP protocol can be analyzed by several molecular methods including end- point RT-PCR and quantitative RT-PCR (if binding targets of the RBP are known), microarray, or deep sequencing methods (Figure 3). Given RNA targets of a known sequence, gene specific primers can be designed that allow validation (and quantification) of the immunoprecipitated RNA. Once successful RIP is confirmed, further interrogation of the population of RNAs in an immunoprecipitation may be pursued by population-based methods such as comparative microarray hybridization of resulting cDNAs or by deep sequencing of molecularly adapted products from the RIP. Presented below are illustrative methods for performing end-point or real time quantitative measurement or NGS analysis of RIP experiments.

Figure 3.RIP RNAs analyses by qRT-PCR
A. RIP Lysate prepared from HeLa cells (2x107cell equivalents per IP) were subjected to immunoprecipitation using 5 µg of either a normal mouse IgG or the anti-PABPC1 antibody and the Magna RIP Kit (Product No. 17-700). Successful immunoprecipitation of PABPC1-associated RNA was verified by qRT-PCR using RIP Primers, ACTB.
B.RIP Lysate prepared from HeLa cells (0.5 × 106 or 1.7 × 106 cell equivalents per IP) were immunoprecipated using 2.5 μg of either a normal Rabbit IgG or Anti-HUR antibody and the Imprint RNA Immunoprecipitation Kit (RIP). Immunoprecipitation of HUR-target and non-target associated RNA was validated by qPCR using Control Primers Actin B and JUN.
C. LincSFPQ associated with the PRC2 complex (EZH2 and SUZ12) and U1snRNA (negative), in HeLa cells were analyzed by qRT-PCR with Magna Nuclear RIP cross-linked (Product No. 17-10520). The signal to noise ratio performances were compared with a competitor kit.
D and E. Magna Nuclear RIP Assay with as few as 5,000 (Cross-linked, Product No. 17-10520) and 100,000 (Native, Product No. 17-10522) Cells. Sensitivity data showing that crosslink protocol NEAT1 enrichment was shown with 5,000 HeLa Cells while with native methods required 100,000 cells (C). Signa to noise ration data showing that native protocol overall positive to negative ratio was higher (D).
F. MeRIP (Cat. #: 17-10499) was performed using total RNA from HEK293 cells. Purified RNA was then analyzed by RT-qPCR using Positive Control Primers (MeRIP Primers Human EEF1A1 Positive, Part # CS220017) and Negative Control Primers (MeRIP Primers Human EEF1A1 Negative, Product No. CS220018).
G. Left panel: successful retrieval of methylated RNA by MeRIPTMm6A Assay with mRNA from various cells. MeRIP (Product No. 17-10499) was performed using mRNA from xeno-free human foreskin fibroblast cells (HFF, Product No. SCC058), UM-SCC-104 human head & neck squamous carcinoma cells (Product No. SCC072), and human embryonic stem cells (H9). Purified RNA was then analyzed by RT-qPCR using Positive Control Primers (MeRIP Primers Human EEF1A1 Positive, Product No. CS220017) and Negative Control Primers (MeRIP Primers Human EEF1A1 Negative, Product No. CS220018). Middle and Right panels: Analysis of m6A RNA Levels of Reprograming Genes in Mouse and Human ES cells. MeRIP (Product No. 17-10499) was performed using mRNA from PluriStem 129/S6 murine ES cells (Product No. SCR012) and H9 human ES cells. Purified RNA was then analyzed by RT-qPCR using MeRIP m6A peak primers for Myc, Sox2, Nanog, Klf4, and Oct4. PCR primer sequences for mouse reprograming genes were provided by Howard Chang (Stanford University). Human PCR primers were designed from published MeRIP-Seq data. One step RT-qPCR was performed with Bio-Rad qPCR machine and iTaq ™ Universal One-Step Kits (Bio-Rad). Percent input recovery was calculated by standard curve drawn with 0.1% input sample or delta delta Ct methods.
H. ChIRP (Product No. 17-10494) was performed using HeLa cell lysate and either Magna ChIRP TERC lncRNA Probe Set even odd (Product No. 03-309) or NEAT1 lncRNA Probe Set even odd (Product No. 03-308)or Magna ChIRP Negative Control Probe Set (LacZ) (Product No. 03-307). Purified RNA was then analyzed by qRT-PCR using RNA Positive Control Primers (TERC or NEAT1) and RNA Negative Control Primers (GAPDH).
RIP RNAs analyzed by Next Generation Sequencing (RIP-seq)

Figure 4.RIP-Seq analysis was performed with EZH2 and SUZ12 in HeLa cells using either Magna nuclear native RIP kit (Product No. 17-10522) or Magna nuclear cross-linked RIP kit (Product No. 17-10520). The data showed enriched signals at NEAT1 (A, top panel) and HOTAIR lincRNA transcripts (B. middle and bottom panels).

Figure 5.Next Generation Sequencing Analysis of DNA pull-down using Magna ChIRP kit
ChIRP was performed on a HeLa cell lysate. Biotinylated capture oligo (Magna ChIRP™ NEAT1 lncRNA Probe Set (odd and even pools) (Product No. 03-308)) and negative control probes (Magna ChIRP™ Negative Control Probe Set (LacZ) Product No. 03-307) were used for pull down reactions. The sequence libraries were sequenced on HiSeq instrument (Illumina). The sequence reads were aligned to the reference genome (hg19) using Bowtie. (a) Peaks were called separately using data from even
and odd probe sets. Algorithms such as MACS can be used for this purpose. Those in common were considered to be valid peaks. (b) A series of post-alignment processing and filtering steps were carried out using analysis software available from the laboratory of Howard Chang (h/HU/enttp://changlab.stanford.edu/protocols.html). The data showing localization of NEAT1 RNA to the NEAT1 gene region and another example region (c).
RIP FAQs, Troubleshooting Tips and Data Analysis Guide
while conducting RIP experiments, you may encounter some challenges, such as no RNA immunoprecipitation or high background or low signal to noise ratio. The information provided below are to help you gain enough understanding of this technique before you start it; provide tips when the experiments do not work and guide you through the data analysis.
- Whole cell lysate RIP FAQs
- Troubleshooting tips for whole cell lysate RIP
- Troubleshooting tips for Chromatin Isolation by RNA Purification (ChIRP)
- Troubleshooting tips for MeRIP
- RIP-qPCR brief protocol
- RIP-qRT-PCR: Data Analysis Calculation Shell
Which RIP kit is right for you?
Whole cell lysate RIP
Nuclear RIP (cross-linked and Native)
Chromatin Isolation by RNA Purification (ChIRP) basick kits and different lncRNA probe sets
meRIP
To continue reading please sign in or create an account.
Don't Have An Account?