Use of Peptide Libraries to Investigate the Evolutionary Preservation of Antimicrobial Properties of Complement Peptide C3a from Invertebrates to Humans
Mukesh Pasupuleti1, Bjorn Walse2, Emma Andersson Nordahl1, Matthias Morgelin1, Martin Malmsten3, Artur Schmidtchen1
1Department of Clinical Sciences, Lund University, Biomedical Center, Tornavagen 10, SE-221 84 Lund, 2SARomics AB, P.O. Box 724, SE-220 07 Lund, 3Department of Pharmacy, Uppsala University, Sweden
Introduction
Proteins that are essential to the survival of an organism are often structurally or functionally conserved to some extent throughout evolution. An example of an evolutionary old molecule is the C3 complement factor that is found both in modern day humans as well as in the horseshoe crab, Carcinoscorpius rotundicauda, a protostome considered a living fossil originating over 500 million years ago1. The human anaphylatoxin peptide C3a is one of multiple proteolytic fragments released in the C3 step of complement activation2, and this protein is generally conserved in vertebrates. The C3a protein performs not only multiple proinflammatory functions, but also exerts potent antimicrobial effect against Gram-negative and Gram-positive bacteria3. C3a and other antimicrobial peptides play important roles in the immune system by providing a rapid and non-specific response against the invasion of pathogenic microorganisms. Indeed, human4 and animal5,6 models with C3 deficiency have been demonstrated to be more susceptible to bacterial infections.
On the other hand, C3a has been found in animals that lack adaptive immunity, such as the pre-historic C. rotundicauda1and the deuterostome Ciona intestinalis7. Yet these animals have an effective antimicrobial defense against pathogenic microorganisms. So this study is an attempt to determine what is lost and what is conserved in this long evolutionary process. A combination of phylogenetic analyses, amino acid sequence alignments, structural homology alignments, biophysical characterization and functional analyses were used to identify the structural pre-requisites governing antimicrobial activity of C3a from humans back to prehistoric invertebrates such as C. rotundicauda.
Experimental
The experimental strategies involved phylogenetic analyses of C3a, C4a, C5a amino acid sequences to identify and select sequences with greater than 70% structural homologies8. These homologous sequences were used for phylogenetic tree reconstruction9 rooted from human C3a. A series of 20-mer peptides corresponding to various regions of C3a were synthesized using the PEPscreen® Custom Peptide Libraries platform (Sigma). These peptides were screened for antimicrobial activity by using the radial diffusion assay (RDA)10 against the bacterial P. aeruginosa 27.1 and E. faecalis 2374. The secondary structures of peptides in solution were determined using CD-spectroscopy. In addition, computational modeling was used to provide structural information, especially in reference to the recently published structure of the C3 protein.
Finally, the ability of the model peptides to perturb membrane structure was also assessed.
Sequence and Phylogenetic Analysis
Representative C3a peptide from vertebrates and invertebrates showed that the C3a molecules co-localized to a single group (Figure 1), suggesting that the C3a has evolved via multiple changes in the protogene. This is consistent with previous analyses of the evolution of the complement system1,11 and clearly supports a common ancestral protein. On the other hand, C4a and C5a formed separate clades, C5a being most distant from the C3a. This pattern suggests that C5a and C4a likely evolved from C3a and that C5a is the paralog of C4a and C3a1.
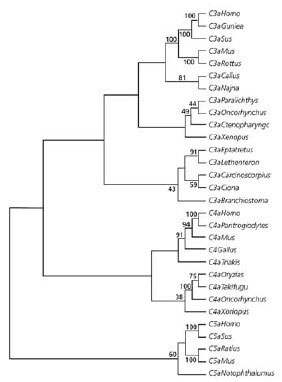
Figure 1.Sequence and Phylogenetic Analysis The evolution of anaphylotoxins. Phylogenetic tree representing C3a, C4a, C5a sequences using the neighbor joining group. Numbers on the branches indicate the reliability of the branch using 1000 bootstrap replications.
Closer examination of this phylogenetic relation revealed several common structural features that are vital for the integrity and stability of the corresponding C3a, C4a, and C5a molecules from various organisms (Figure 2). For example, six disulfide-bonded cysteines are conserved across species and three disulfide bonds appear to be vital in stabilizing the conformation of the internal “core” portion of the molecules. The presence of other highly conserved residues suggests their importance for the structural stability as well as function of C3a.
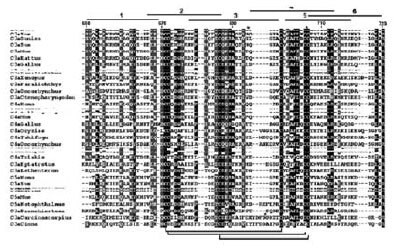
Figure 2.Phylogenetic Relation Analysis Alignment of C3a, C4a, and C5a sequences showing identical and similar amino acid substitutions. The shading represents the degree of conservation at each position in the alignment, taking into account similar physicochemical properties of the residues. The peptide sequences corresponding to regions 1-6 in the human C3a sequence are indicated. Note that this only applies to the C3a sequences. Disulfide bonds are indicated at the bottom. The 20-mer peptide libraries are indicated as.
Structural Modeling
Despite extensive overall sequence discrepancy, a clear similarity is observed between the observed structure of human C3a and the predicted structure of the corresponding C3a peptide from C. rotundicauda (Figure 3A). Both peptides have striking similarity in the four helical regions and in the two first loops located before the cysteines at positions 22 and 36 and positions 17 and 31 in the human and C. rotundicauda peptides, respectively. Another similar feature is the presence of prominent cationic and amphiphatic helical C-terminus, which are also seen in the helical wheel diagrams and the 3-D models (Figure 3B).
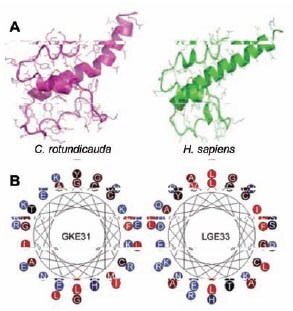
Figure 3. (A) Molecular models. Predicted structures of C. rotundicauda and human C3a proteins. Human C3a is represented by the corresponding part in the crystal structure of human C3 (PDB code 2A73). (B)Helical wheel representation of the C3a-deruived C-terminal peptides GKE31 from C. rotundicauda and LGE33 from humans
Antimicrobial activities
In order to explore the structurefunction relationships of C3a epitopes, overlapping peptide sequences comprising 20 mers (Figure 2) were synthesized and screened for antimicrobial activities against P. aeroginosa, a ubiquitous pathogen found among vertebrates and invertebrates. Most antimicrobial peptides are known to exhibit common minimum levels of cationicity, amphipathicity, and hydrophobicity6. The results clearly showed that peptides with cationic C-termini exhibited significant antimicrobial activity, while peptides with negatively charge C-termini did not show microbial activity (data shown in original paper). Phylogenetic analyses showed that that animals whose C3a peptides exhibited antimicrobial activity belongs to subclades that are different from those animals whose C3a peptide did not have antimicrobial activity. In general, peptides displaying antimicrobial activity had a net charge of +2 to +4 and ~20 to 40% hydrophobic amino acids. The degree of amphipathicity, as judged by the relative hydrophobic moment (mHel) ranged from ~0.2-0.4 and consistent with those observed in many helical peptides exhibiting antimicrobial activity12. The results also confirmed that net charge correlates to antimicrobial activity. These findings are consistent with the evolution of primate cathelicidin, where positive selection affected the number of charges while keeping hydrophobicity and amphipathicity fairly constant12.
Structural and Functional Congruence
Antimicrobial activity assays showed that peptides representing human (LGE33) and C.rotundicauda (LKE31) C3e sequences (Figure 3B) exerted similar antibacterial effects against both Gram-negative P. aeruginosa and E. coli and the Gram positive B. subtilis (Figure 4A). Fluorescence and electron microscopy studies revealed that both peptides caused local perturbations and breaks along P. aeruginosa plasma membranes (data shown in original article), an observation that is very similar to the “classical” human cathelicidin LL-37. Circular dichroism studies showed that both peptides did not have any appreciable structure in aqueous solutions (Figure 4B). However, despite a marked difference in their primary sequences, both peptides exhibited large and almost identical induction of helicity occurs upon interactions of the peptides with E. coli lipopolysaccharide (LPS). Similarly, at 1 μM concentration both peptides induced leakage of liposomes (Figure 4C), reaching 80% maximum leakage within the first 200 seconds (Figure 4D).
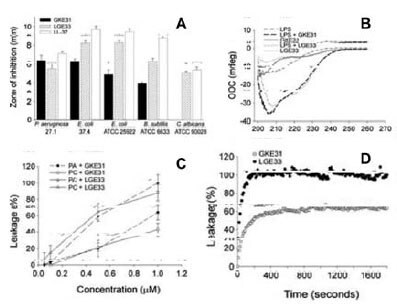
Figure 4.Activities of C3a-derived human (LGE33) and C. rotundicauda (LKE31) peptides. (A) Antibacterial effects based on radial diffusion assay. (B) CD spectrum in Tris buffer (control) and in the presence of LPS. (C) Effects of the peptides on membrane permeabilization based on leakage of carboxyfluorescein from liposomes. (D) Kinetic analysis of liposome permeabilization.
The results therefore indicate that the GKE31 and LGE peptides indeed function like most helical antimicrobial peptides such as LL- 37 by interacting with the lipopolysaccharides, leading to induction of an a-helical conformation, which in turn facilitates membrane interactions, membrane destabilization and, finally, bacterial killing. The fact that the two C3a-derived peptides are separated by as much as over half billion years of evolutionary distance elegantly demonstrates the remarkable structural and functional conservation of this C-terminal region.
Conclusions
Phylogenetic analysis, sequence analyses, structural modeling studies, structural analyses, and antimicrobial assays of peptides from known C3a sequences have together shown the crucial structural determinants governing antimicrobial activity that have been conserved during the evolution of C3a in invertebrates and vertebrates. Thus, regions of the ancient C3a from C. rotundicauda, as well as corresponding parts of human C3a, exhibited helical structures upon binding to bacterial lipopolysaccharides, permeabilised liposomes, and the retention of positive charges in these helices impart antimicrobial activity against Gram-negative and Gram-positive bacteria. Animals whose C3a proteins contain negatively-charged helices lost antimicrobial resistance, and the protein appears to have evolved for a different function, such as into a purely chemotactic and highly spasmogenic molecule. The evolutionary process has positively selected many structurally diverse antimicrobial peptides that enable the organism to adapt to the microbial environment, as well as introduction of novel biological functions. Finally, organisms continuously evolve to generate molecules, such as C3a, that provide a robust and structurally intact defense mechanism against the selection forces.
References
To continue reading please sign in or create an account.
Don't Have An Account?