ChIP Tested Antibodies
Chromatin structure plays a fundamental role in regulating processes that take place on the DNA, such as transcription, replication, and recombination, all of which occur on nucleosomal templates. Therefore, mapping the position of histones selectively modified and histone variants or non-histone components of chromatin in specific DNA sequences, can provide valuable insights into how these proteins (and their modifications) function in a chromatin context.
Chromatin immunoprecipitation (ChIP) is a powerful tool for identifying proteins, including histone proteins and non-histone proteins, associated with specific regions of the genome by using specific antibodies that recognize a specific protein or a specific modification of a protein. The method is based on the principle that formaldehyde reacts with primary amines located on amino acids and the bases on DNA or RNA molecules, forming a covalent crosslink between the specific protein to the DNA on which they are situated.
Once the DNA binding proteins are crosslinked, the cells are lysed and the crude cell extracts are sonicated to shear the DNA to a smaller size. The protein: DNA complex is immunoprecipitated using an antibody against the protein of interest. The DNA protein cross-links are reversed by heating and the proteins removed by treatment with proteinase K. The DNA portion of the complex is then purified and identified by PCR using specific primers to the suspected binding region.
Anti-Acetylated Proteins antibody produced in rabbit
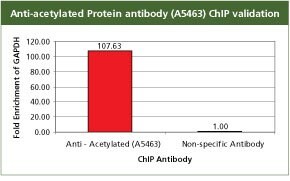
For validation the product A5463, Anti-Acetylated Proteins antibody produced in rabbit with the CHP1 kit. GAPDH is a ubiquitously transcribed gene, hence the acetylation transcription mark is enriched on its promoter in HeLa cells. For each reaction, 2.5 µg of antibody and 250,000 cells were used followed by qPCR analysis. All the buffers used in the protocol were made with Sigma products. PCR was done using S9939, the SYBR JumpStart ReadyMix.
GAPDH Forward 5' CAA TTC CCC ATC TCA GTC GT 3’
GAPDH Reverse 5' TAG TAG CCG GGC CCT ACT TT 3'
Non-specific antibody Anti mouse IgG (I8765)
For the PCR reaction, used 2 µl of template DNA from a 30 µl chip reaction; PCR were run in 25 µl total volume.
Cycling conditions started with a
95 °C denaturation for 3 min followed by forty cycles of
95 °C denaturation for 20 sec,
60 °C annealing for 20 sec,
72 °C extension for 20 sec,
82 °C fluorescence read for 20 sec.
This four-step protocol removes any contribution of primer associated products in the fluorescent read.
Monoclonal Anti-acetyl-Histone H3 (Ac-Lys9) Antibody Produced in Mouse
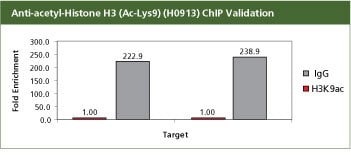
For validation the product H0913, Monoclonal Anti-acetyl-Histone H3 (Ac-Lys9) antibody produced in mouse with the CHP1 kit. H3K9ac is an activation mark and hence should be associated with the expressed genes BMP7 and ELF3S10. Used with 10 million SW480 cells in a traditional Staphylococcus aureus ChIP method. In SW480 cells the ChIP data shows that the H3K9ac mark is enriched with promoters BMP7 and ELF3S10. All the buffers used in the protocol were made with Sigma products. PCR was done using S9939, the SYBR JumpStart ReadyMix.
BMP7 Forward 5' AGG ATC CCT CCT TCC CAG TA 3'
BMP7 Reverse 5' GGG CTT CTC CTA CCC CTA CA 3'
EIF3S10 Forward 5' TGA CGA CAC TCC TTT CCG TG 3'
EIF3S10 Reverse 5' CCT GGG AAT CGC CTG TCT TG 3'
For the PCR reaction used 1.5 µl of template DNA from a 30 µl chip reaction; PCR were run in 25 µl total volume
Cycling conditions for PCR started with
95 °C denaturation for 4 min followed by thirty-three cycles of
94 °C denaturation for 30 sec,
60 °C annealing for 30 sec,
72 °C extension for 30 sec,
PCR reaction was concluded with a one-minute, 72 °C extension step
Monoclonal Anti-Histone Deacetylase 2 (HDAC2) Antibody Produced in Mouse
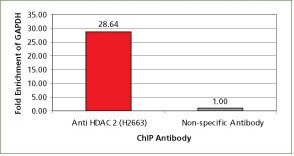
For validation the product H2663, Monoclonal Anti-Histone Deacetylase 2 (HDAC2) antibody produced in mouse with the CHP1 kit. The ubiquitous housekeeping gene, GAPDH, is enriched with HDAC2 in HeLa cells. For each reaction, 2.5 µg of antibody and 250,000 cells were used followed by qPCR analysis. All the buffers used in the protocol were made with Sigma products. PCR was done using S9939, the SYBR JumpStart ReadyMix.
GAPDH Forward 5' CAA TTC CCC ATC TCA GTC GT 3'
GAPDH Reverse 5' TAG TAG CCG GGC CCT ACT TT 3'
Non-specific antibody Anti mouse IgG (I8765)
For the PCR reaction used 2 µl of template DNA from a 30 µl chip reaction; PCR were run in 25 µl total volume
Cycling conditions started with a
95 °C denaturation for 3 min followed by forty cycles of
95 °C denaturation for 20 sec,
60 °C annealing for 20 sec,
72 °C extension for 20 sec,
82 °C fluorescence read for 20 sec.
This four-step protocol removes any contribution of primer associated products in the fluorescent read.
Anti-methyl-Histone H3 (Me-Lys9) Antibody Produced in Rabbit
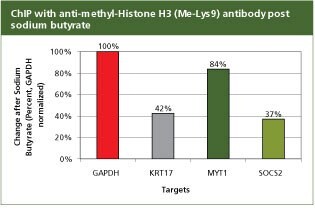
For validation the product H7162, Anti-methyl-Histone H3 (Me-Lys9) antibody produced in rabbit with the CHP1 kit. To analyze promoter regions for GAPDH, KRT17, MYT1, and SOCS2 in SW480 cells following a 24 hour treatment with 5 mM Sodium Butyrate (B5887), a histone deactylase (HDAC) inhibitor. Values were normalized to GAPDH levels. 2 µg of antibody H3K9me (H7162) and 250,000 cells were used per reaction. A post treatment decrease in the association of KRT17, MYT1, and SOCS2 regions with H3K9me is expected as they are target genes of the polycomb repressor complex which requires HDAC activity to represses/silence the transcription of its target genes.
GAPDH Forward 5' CAA TTC CCC ATC TCA GTC GT 3'
GAPDH Reverse 5' TAG TAG CCG GGC CCT ACT TT 3'
KRT17 Forward 5' TTG GGG TAC AGA AGG GTG AG 3'
KRT17 Reverse 5' TCC CCA GGT TTA CAC TCC AG 3'
MYT1 Forward 5' AGG CAC CTT CTG TTG GCC GA 3'
MYT1 Reverse 5' AGG CAG CTG CCT CCC GTA CA 3'
SOCS2 Forward 5' GAG TTC GAG ACC AGC CTG AC 3'
SOCS2 Reverse 5' GAC CAA GTC TCC CTC TGT CG 3'
For the PCR reaction used 2 µl of template DNA from a 30 µl chip reaction; PCR were run in 25 µl total volume
Cycling conditions started with a
95 °C denaturation for 3 min followed by forty cycles of
95 °C denaturation for 20 sec,
60 °C annealing for 20 sec,
72 °C extension for 20 sec,
82 °C fluorescence read for 20 sec.
This four-step protocol removes any contribution of primer associated products in the fluorescent read.
Anti-acetyl-Histone H1.4 (Ac-Lys26) Antibody Produced in Rabbit
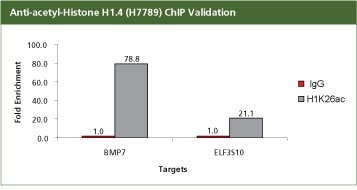
For validation the product H7789, Anti-acetyl-Histone H1.4 (Ac-Lys26) antibody produced in rabbit with the CHP1 kit. 2.8 µg of antibody H1K26ac (H7789) was used with 400,000 SW480 cells. Fold Enrichment was calculated using anti rabit IgG as a non-specific antibody. In SW480 cells the ChIP data shows that the H1K26ac mark is enriched with BMP7 and ELF3S10 promoters, which are not repressed. H1K26ac is an activation mark and hence should be associated with expressed genes BMP7 and ELF3S10. All the buffers used in the protocol were made with Sigma products. PCR was done using S9939, the SYBR JumpStart ReadyMix.
BMP7 Forward 5' AGG ATC CCT CCT TCC CAG TA 3'
BMP7 Reverse 5' GGG CTT CTC CTA CCC CTA CA 3'
EIF3S10 Forward 5' TGA CGA CAC TCC TTT CCG TG 3'
EIF3S10 Reverse 5' CCT GGG AAT CGC CTG TCT TG 3'
For the PCR reaction used 2 µl of template DNA from a 30µl chip reaction; PCR were run in 25 µl total volume
Cycling conditions started with a
95 °C denaturation for 3 min followed by forty cycles of
95 °C denaturation for 20 sec,
60 °C annealing for 20 sec,
72 °C extension for 20 sec
82 °C fluorescence read for 20 sec.
This four-step protocol removes any contribution of primer associated products in the fluorescent read.
Anti-acetyl- & phospho-Histone H3 (Ac-Lys9, pSer10) Antibody Produced in Rabbit
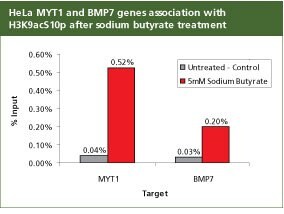
For validation the product H9161, Anti-acetyl- & phospho-Histone H3 (Ac-Lys9, pSer10) antibody produced in rabbit with the CHP1 kit was used to analyze promoter regions for Myelin transcription factor 1 (MYT1) and Bone Morphogenetic Protein 7 (BMP7) in HeLa cells following 24 hours with 5 mM Sodium Butyrate (B5887). The enrichment of MYT1 and BMP7 genes associated with H3K9acS10p is expected following treatment with the histone deactylase inhibitor (HDAC), sodium butyrate. 1 µg of antibody H3K9acS10p (H9161) was used with 250,000 cells. All the buffers used in the protocol were made with Sigma products. PCR was done using S9939, the SYBR JumpStart ReadyMix.
MYT1 Forward 5’ AGG CAC CTT CTG TTG GCC GA 3’
MYT1 Reverse 5’ AGG CAG CTG CCT CCC GTA CA 3’
BMP7 Forward 5’ AGG ATC CCT CCT TCC CAG TA 3’
BMP7 Reverse 5’ GGG CTT CTC CTA CCC CTA CA 3’
For the PCR reaction used 2 µl of template DNA from a 30 µl chip reaction; PCR were run in 25 µl total volume
Cycling conditions started with a
95 °C denaturation for 3 min followed by forty cycles of
95 °C denaturation for 20 sec,
60 °C annealing for 20 sec,
72 °C extension for 20 sec
82 °C fluorescence read for 20 sec.
This four-step protocol removes any contribution of primer associated products in the fluorescent read.
Anti-acetyl-Histone H3 (Ac-Lys9) Antibody Produced in Rabbit
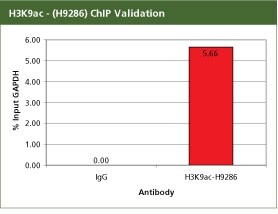
For validation the product H9286, Anti-acetyl-Histone H3 (Ac-Lys9) antibody produced in rabbit with the CHP1 kit. For each reaction, 2 µg of Anti-acetyl-Histone H3 (Ac-Lys9) (H3K9ac) (H9286) antibody was used with 10 million SW480 cells in a traditional Staphylococcus aureus ChIP method. Fold enrichment was calculated using anti-mouse IgG as a non-specific antibody. H3K9ac is an activation mark and hence should be associated with the promoter of the ubiquitously transcribed GAPDH gene. In SW480 cells this ChIP data shows that the H3K9ac mark is enriched with GAPDH promoter. All the buffers used in the protocol were made with Sigma products. PCR was done using S9939, the SYBR JumpStart ReadyMix.
GAPDH Forward 5' CAA TTC CCC ATC TCA GTC GT 3'
GAPDH Reverse 5' TAG TAG CCG GGC CCT ACT TT 3'
For the PCR reaction used 1.5 µl of template DNA from a 30 µl chip reaction; PCR were run in 25 µl total volume
Cycling conditions for PCR started with
95 °C denaturation for 4 min followed by thirty three cycles of
94 °C denaturation for 30 sec,
60 °C annealing for 30 sec,
72 °C extension for 30 sec
PCR reaction was concluded with a one minute, 72 °C extension step
Anti-RbAp46, C-terminal Antibody Produced in Rabbit
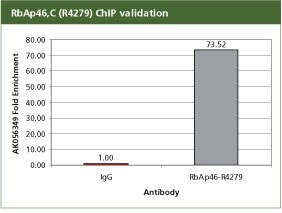
For validation the product R4279, Anti-RbAp46, C-terminal antibody produced in rabbit with the CHP1 kit. For each reaction, 2 µg of anti-RbAp46 (R4279) antibody was used with 10 million SW480 cells in a traditional Staphylococcus aureus ChIP method. Fold enrichment was calculated using anti-mouse IgG as a non-specific antibodyRbAp46 is a component of the polycomb repressor complex (PRC), hence AK056349 is enriched on the known PRC target. All the buffers used in the protocol were made with Sigma products. PCR was done using S9939, the SYBR JumpStart ReadyMix.
AK056349 Forward 5' AGG GCA ACT GGG CAT GCT CA 3'
AK056349 Reverse 5' CAC GCC CTT TTC CCT CTG TC 3'
For the PCR reaction used 1.5 µl of template DNA from a 30 µl chip reaction; PCR were run in 25 µl total volume
Cycling conditions for PCR started with
95 °C denaturation for 4 min followed by thirty three cycles of
94 °C denaturation for 30 sec,
60 °C annealing for 30 sec,
72 °C extension for 30 sec
PCR reaction was concluded with a one-minute, 72 °C extension step
References
To continue reading please sign in or create an account.
Don't Have An Account?