MVM Resistance Through Genetic Engineering
Contamination in the manufacturing process can be a rare but catastrophic event costing a company millions per contamination in clean-up costs along. This does not include any loss of revenue due to missed product sales or the impact to the patient population due to loss of drug supply. Even with the use of animal origin free and animal component free systems, there have been recorded contaminations of CHO cells by Minute Virus of Mice (MVM) also called Mouse Minute Virus (MMV).
As part of the CHOZN brand promise to enable the development of better biologics and safer manufacturing processes, we have identified a gene target (SLC35A1) that upon elimination of expression by gene editing, results in a CHO cell line that is resistant to infection by MVM. In addition, protein titer is not affected by this modification.
- The Science behind MVM Resistance
- Zinc Finger Nuclease (ZFN) Targeting SLC35A1
- Validation of MVM Resistance
- Growth and Productivity in SLC35A1 Knockout cells
- Case Study
- Conclusion
The Science behind MVM Resistance
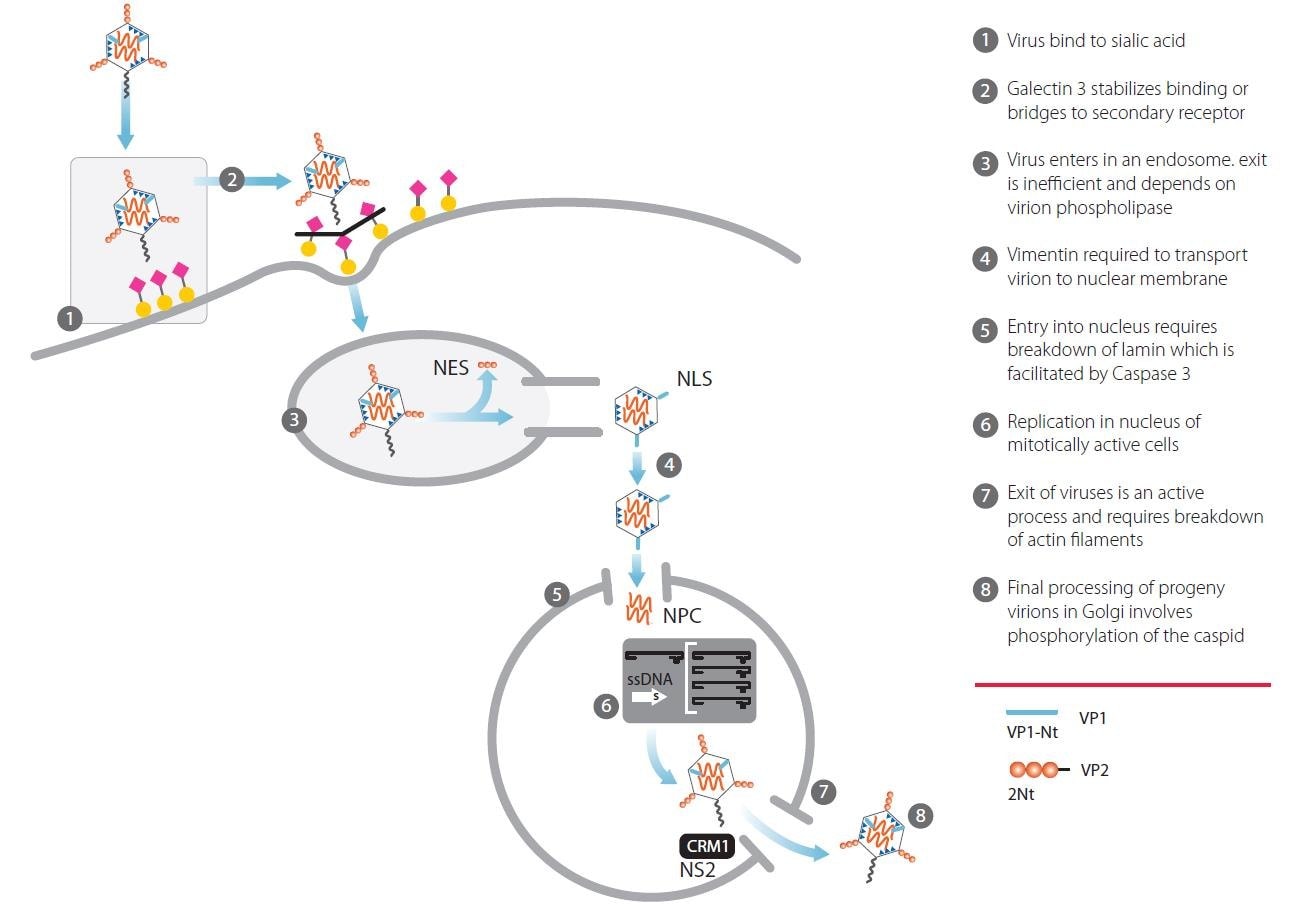
Figure 1.Infection and replication of Minute Virus of Mice in a host cell line
Attachment to a cell surface receptor is a key first step in the infection cycle for many viruses. In 2006, a glycan array approach used to study MVM viral capsid binding (Nam et al., Sept 2006, JBC 281, no 35, p25670) reported that viral capsids derived from MVMp bound to specific glycan structures with terminal sialic acid.
The protein produced by SLC35A1 is responsible for transporting sialic acid into the Golgi. Knocking out function of this gene in a cell results in asialylated glycan structures, thus eliminating the ability of MVM to bind to and enter the cell.

Figure 2.Theoretical phenotypes of N-linked and O-linked sialic acid as a result of SLC35A1 knockout.
Zinc Finger Nuclease (ZFN) Targeting SLC35A1
ZFN gene editing reagents were designed to target SLC35A1. ZFNs are a class of engineered DNA-binding proteins that facilitate targeted editing of the genome by creating double-strand breaks in DNA at user-specified locations. An active ZFN reagent was identified targeting SLC35A1 in CHO cells.
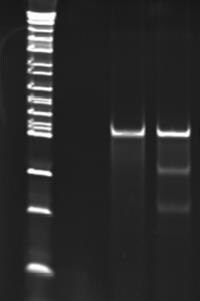
Figure 3.Results of CEL I mismatch detection assay. The two lower bands in lane 2 indicate the successful targeting and DNA cutting ability of the ZFN.
Lane 1) Mock transfection sample
Lane 2) Sample transfected with ZFN RNA showing
approximately 14% modification frequency
A single cell clone was isolated from the ZFN transfected population and genotyped to confirm modification of SLC35A1. A phenotypic assay using Mal II lectin which binds specifically to surface sialic acid was run verifying the generation of an asialylated cell line.

Wildtype CHO – Fluorescent

SLC35A1 KO CHO – Fluorescent

Wildtype CHO – Brightfield
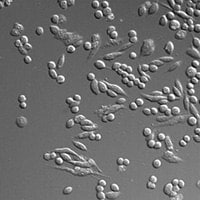
SLC35A1 KO CHO – Brightfield
Figure 4. Wildtype and knockout (KO) cells were incubated with MAL II-biotin, washed and then incubated with Streptavidin-Alexa-647. Cells were washed and observed under a microscope
Validation of MVM Resistance
Both wildtype and SLC35A1 knockout cells were exposed to MVM. Immediate loss of viability and lack of propagation was observed in the exposed wildtype cells; however, growth was not affected by exposure to MVM in the SLC35A1 knockout cells indicating the wildtype cells are susceptible to infection while the SLC35A1 knockout cells are resistant.

Graph 1.Wildtype and SLC35A1 knockout cells infected with MVM at an MOI of 1.0. Viable cell density determined through day 8 post infection.
MVM In-Vitro Challenge Assay - Single Cycle Replication
Wildtype and SLC35A1 knockout cells were incubated with MVM for 3 hours and then washed to remove non-specifically bound virus. Samples were then collected at time=0 hours and time=21 hours and analyzed by qPCR using a probe against the MVM NS2 gene. Time 0 indicates levels of viral attachment. Time 21 indicates levels of internalization and single cycle replication.

Graph 2.Wildtype and SLC35A1 knockout cells exposed to MVM. MVM replication is abrogated in SLC35A1 knockout cell line.
MVM In-Vitro Challenge Assay - Extended Culture Replication
Wildtype and SLC35A1 knockout cells were incubated with MVM for 3 hours and then washed to remove non-specifically bound virus. Samples were then collected at time=0 hours and every 24 hours up to 96 hours and analyzed by qPCR using a probe against the MVM NS2 gene. Viral genome copies increase with extended time in the wildtype cells. In the SLC35A1 knockout cells, the viral genome copy number does not rise above inoculum levels indicating complete resistance to MVM infection.

Graph 3.Wildtype and SLC35A1 knockout cells exposed to MVM. MVM replication is abrogated in SLC35A1 knockout cell line.
Growth and Productivity in SLC35A1 Knockout cells
Multiple SLC35A1 knockout clones were evaluated for growth compared to wildtype cells. Viable cell density was measured over a 13 day assay. The SLC35A1 knockout clones showed normal clonal variability in growth patterns compared to the wildtype cells.

Graph 4.Viable cell density seen in wildtype and SLC35A1 knockout clones over a 13 day fed-batch assay.
The top four clones were chosen based on growth profiles. These clones, along with the wildtype cells, were transfected with a vector expressing a model IgG1 molecule. The transfected populations were plated into pools in 96-wells plates and 7-day static productivity was measured. The four clones demonstrated a range in titers, that when compared to the wildtype titer, indicated the genetic modification did not have a detrimental effect on growth or productivity.
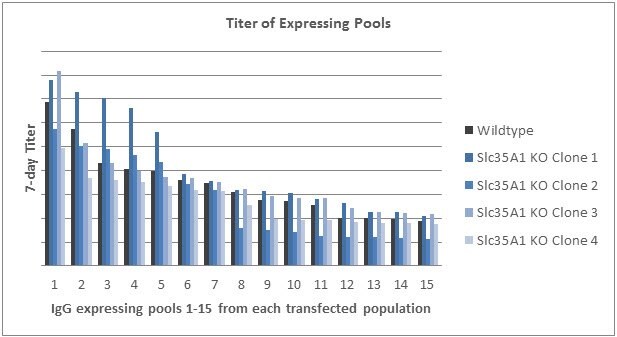
Graph 5.Titer obtained from fifteen pools isolated from each of the transfected populations. Pools from each population are ordered from highest titer to lowest.
Case Study – SLC35A1 Knockout in an Expressing Production Line
Growth and Productivity
Both wildtype and SLC35A1 knockout clones were isolated from the transfected population. Viability, viable cell density and productivity were measured. It was determined that the genetic modification did not impact these parameters. A range of titer and VCD were obtained in both knockout and wildtype clones.

Graph 6.Viability and viable cell density of both SLC35A1 knockout and wildtype clones were measured over the course of a 9 day assay. A range of viable cell densities across wildtype and knockout clones indicate that this genetic modification did not impact this parameter.
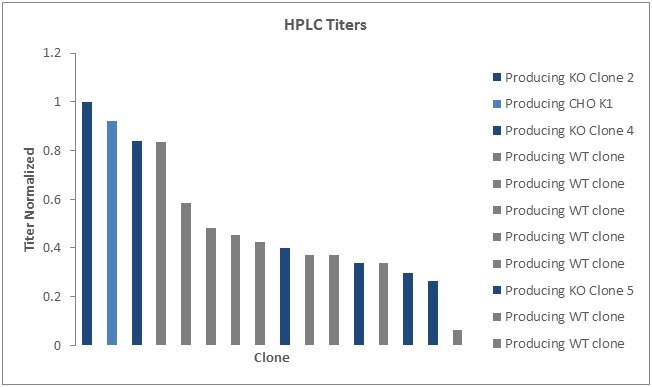
Graph 7.Maximum titer of both SLC35A1 knockout and wildtype clones were obtained. A range of peak titers across wildtype and knockout clones indicate that this genetic modification did not impact this parameter.
MVM In-Vitro Challenge Assay - Single Cycle Replication from BioReliance
Select wildtype producing host cell lines and SLC35A1 knockout clones were incubated with MVM for 3 hours and then washed to remove non-specifically bound virus. Samples were then collected at time=0 hours and time=21 hours and analyzed by qPCR using a probe against the MVM NS2 gene. Time 0 indicates levels of viral attachment. Time 21 indicates levels of internalization and single cycle replication.
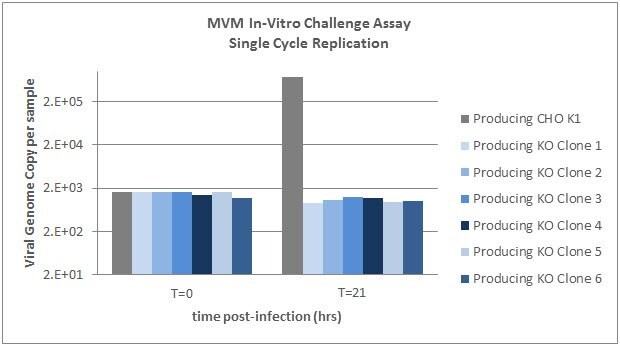
Graph 8.Six IgG producing SLC35A1 knockout clones as well as the wildtype producing cells were exposed to MVM. MVM replication is abrogated in SLC35A1 knockout cell lines.
MVM In-Vitro Challenge Assay - Extended Culture Replication
IgG producing SLC35A1 knockout cells as well as wildtype producing cells were incubated with MVM for 3 hours and then washed to remove non-specifically bound virus. Samples were then collected at time=0 hours and every 24 hours up to 96 hours and analyzed by qPCR using a probe against the MVM NS2 gene. Viral genome copies increase with extended time in the wildtype cells. In the SLC35A1 knockout cells, the viral genome copy number does not rise above inoculum levels indicating complete resistance to MVM infection.
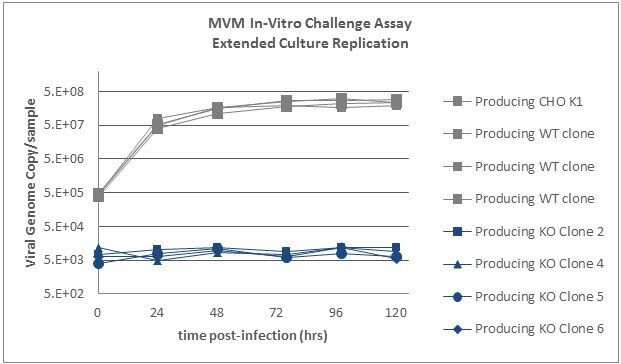
Graph 9.Four IgG producing SLC35A1 knockout clones as well four IgG producing wildtype clones and the parental wildtype producing cells were exposed to MVM. MVM replication is abrogated in SLC35A1 knockout cell lines.
Conclusion
We have identified a gene, SLC35A1, that upon elimination of expression via gene editing technology, confers resistance to MVM in the host cell line. We hypothesize this resistance is due to a lack of MVM internalization into the cells due to the loss of sialic acid at the surface of the cell. We have confirmed this resistance via MVM challenge assays run on SLC35A1 wildtype and SLC35A1 knockout cells. Both single cycle and extended culture replication challenge assays show infection of the wildtype cells and complete resistance to MVM infection in SLC35A1 knockout cells. We have seen no detrimental impact on growth, viability or productivity due to this genetic modification.
To continue reading please sign in or create an account.
Don't Have An Account?