CRISPR/Cas-GFP Vectors for Rapid Expression Verification and Enrichment of Genome Edited Cells
Background
In many genome editing experiments involving ZFNs and CRISPR/Cas nucleases, the first challenge is achieving successful delivery of plasmids and subsequent expression of the encoded nucleases. While CEL-I assays, T7E1 assays, or deep sequencing are ultimately needed to quantify nuclease-induced insertions and deletions (indels), experimental troubleshooting can proceed faster by implementing a method that rapidly reports activity of the nuclease expression cassette. We have developed a Cas9-GFP expression cassette which expedites genome editing workflows from initial nuclease screening to the final stages of single cell cloning. This vector also includes a U6-gRNA cassette creating a single vector system for CRISPR/Cas delivery and expression (Figure 1).
Some cell types and/or genomic locations are more difficult to target with nucleases than others. For situations where repeated trials of CRISPR transfection and testing fail to achieve high level cutting, methods for enrichment of edited cells may be useful. Using our single-vector system, the co-expression of Cas9 and GFP from the same mRNA creates the possibility of enriching cell populations for desired genome edits via fluorescence activated cell sorting (FACS). By being able to detect and enrich with the GFP using FACS, it significantly reduces labor and cost associated with single cell cloning and genotyping in genome editing applications. The following data set illustrates how our single vector system can be used for expression monitoring and FACS enrichment of low and high level cutting.

Figure 1. Schematic of the CRISPR/Cas-GFP vector.
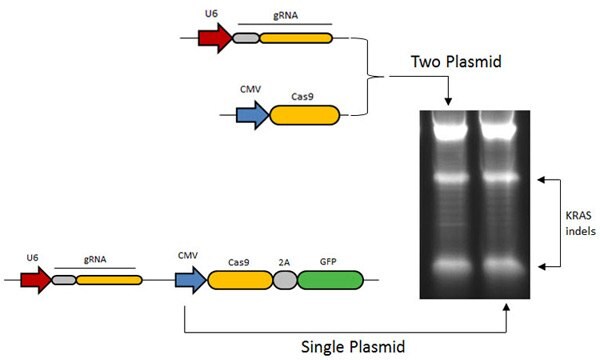
Figure 2. In most current CRISPR applications, gRNA and Cas9 are expressed from two seperate vectors. A gRNA design for the human KRAS locus was shown to retain high level induction of indels in human K562 cells when converted to single vector format. Single plasmid GFP format ensures that all required CRISPR/Cas components (e.g. Cas9 and gRNA coding sequences) are effectively delivered to GFP positive cells.

Figure 3. The delivery of Cas9-GFP expression cassettes can easily be monitored by microscopy or FACS . This particular vector uses the CMV promoter to drive expression of the Cas9-GFP cassette, and while CMV is useful for a broad variety of commonly used cell types, it is not universally active. For example, previous ZFN projects have shown that the CMV promoter is not highly active in HeLa cells (internal, unpublished data). To circumvent this problem in HeLa, ZFN mRNA has been used as an effective alternative to transiently express ZFNs for genome editing applications. If GFP expression and nuclease indel activity cannot be observed in a particular cell type despite repeated attempts, using Cas9-GFP mRNA maybe a good option to circumvent promoter / cell-type incompatibilities. The single vector system described here contains a T7 promoter for in vitro transcription of Cas9-GFP mRNA (Figure 1).

Figure 4. When cell fractions were divided into low, medium, and high pools based on GFP expression as in Figure 3, corresponding increases in indel activity were observed. For a gRNA targeting the KRAS locus, a 4-fold increase in indel activity was observed when comparing the unsorted population vs. the top 2% of cells with the highest GFP expression. Not all targeted gRNA designs produce detectable indel activity when initial nuclease screens are done against gene targets, and current gRNA design rules fail to predict activity based on sequence content or genomic context. We took a gRNA design for CCR5 which initially failed to produce detectable indels and sorted it into low, medium, and high GFP fractions. Indel activity could be detected in the medium and high GFP fractions. This technique can be very useful to rescue the activity of a gRNA design that has a cut site which is located at a position that is optimal for compatibility with a donor DNA (for example, near a disease SNP).
Conclusions
The Cas9-GFP expression cassette provides high level genome editing activity, rapid expression detection, and an option for enriching and rescuing gRNA designs that initially fail to produce detectable indel activity. By using our single plasmid design, it ensures that all GFP positive cells also have the required CRISPR/Cas components. The GFP reporter allows for quick detection of transfection efficiency saving time and cost associated with downstream expression quanification assays. This also allows for rapid troubleshooting of plasmid delivery and expression problems associated with particular cell types. Our CRISPR/Cas technology offering provides a quick and affordable solution to targeted genome editing, allowing access to genome editing to more labs than ever.
To continue reading please sign in or create an account.
Don't Have An Account?