KCQS03
KiCqStart® SYBR® Green qPCR ReadyMix™
iQ™, with fluorescein for Bio-Rad systems
About This Item
Recommended Products
form
liquid
usage
sufficient for 1250 reactions
sufficient for 250 reactions
sufficient for 5000 reactions
feature
dNTPs included
hotstart
storage condition
protect from light
technique(s)
qPCR: suitable
color
colorless
input
purified DNA
compatibility
for use with Bio-Rad MyiQ
for use with Bio-Rad iCycler iQ
for use with Bio-Rad iQ 5
detection method
SYBR® Green
shipped in
dry ice
storage temp.
−20°C
Looking for similar products? Visit Product Comparison Guide
General description
Highly specific amplification is crucial to successful qPCR with SYBR Green I dye technology because this dye binds to and detects any dsDNA generated during amplification. Hot-Start Taq DNA polymerase is antibody mediated to be inactive prior to the initial PCR denaturation step.
Application
Different real-time PCR systems employ different strategies for the normalization of fluorescent signals and correction of well-to-well optical variations. It is critical to match the appropriate qPCR reagent to your specific instrument. KiCqStart SYBR Green qPCR ReadyMix, iQ contains fluorescein for experimental plate well factor collection on iCycler iQ real-time detection systems or the MyiQ detection system.
- Gene expression
- DNA quantification
- CHiP
Features and Benefits
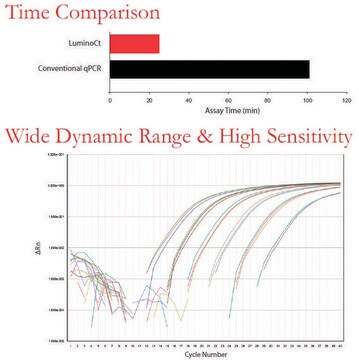
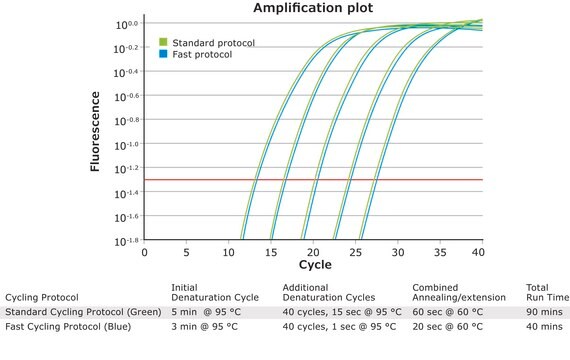
- Assay results in as little as 33 minutes
- Highly efficient and sensitive real-time PCR results
- Little/no optimization required
Components
packaging:
250 reactions* = 2 X 1.25 mL tubes
1250 reactions* = 10 X 1.25 mL tubes
5000 reactions* = 1 X 50 mL tube
*number of reactions based on a 20uL volume
Other Notes
KiCqStart SYBR Green qPCR ReadyMix is stable for 1 year when stored in a constant temperature freezer at -20°C, protected from light. For convenience, it may be stored unfrozen at +2 to +8°C for up to 6 months. After thawing, mix thoroughly before using. Repeated freezing and thawing of the product is not recommended. However, the product demonstrated no loss of performance after 20 freeze-thaw cycles or 2 months at +20°C.
Legal Information
recommended
related product
Storage Class Code
12 - Non Combustible Liquids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
After a traditional PCR has been completed, the PCR/qPCR data analysis is conducted by resolution through an agarose gel or, more recently, through a capillary.
PCR assay guide navigates you through primer validation and other assay optimization factors to ensure high sensitivity and specificity for optimum DNA/ RNA quantification.
Real-time polymerase chain reaction allows researchers to estimate the quantity of starting material in a sample. It has a much wider dynamic range of analysis than conventional PCR
Protocols
Quantitative PCR protocol using SYBR Green reagents. Procedure supports most qPCR instruments.
Analysis of gene expression data requires a stable reference or loading control. This reference is usually one or more reference genes.
Gradient PCR for assay optimization is to determine the optimum annealing temperature (Ta) of the primers by testing identical reactions containing a fixed primer concentration, across a range of annealing temperatures.
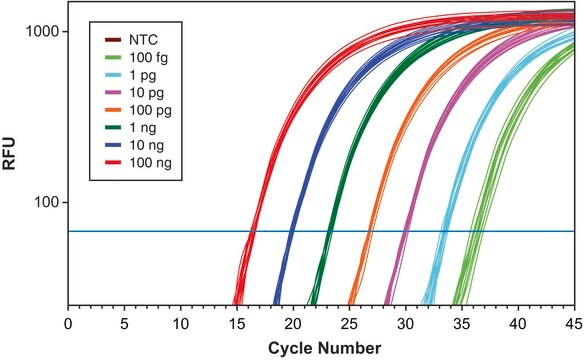
Once an assay has been optimized, it is important to verify the reaction efficiency. This information is important when reporting and comparing assays. In this example protocol, the assay efficiency is compared over a wide and narrow dynamic range of cDNA concentrations.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service