Prestige Antibodies® in Immunohistochemistry
IHC antibodies and the Human Protein Atlas
Immunohistochemistry (IHC) is the most widely used technique in histopathological diagnosis and research for detection of proteins in tissues and cells. Today, IHC can be applied in a high-throughput fashion for studying proteins using Tissue Microarrays (TMAs). In the Human Protein Atlas project, Prestige Antibodies® have been used to analyze all human proteins using IHC and TMAs.1,2 All resulting tissue and cell images are publicly available on the Human Protein Atlas web portal.3,4 In total, more than 500 high resolution IHC images from human tissue samples are presented for each Prestige Antibody®. Each year protein expression and localization data of ~2,500 new proteins were added to the portal. By the end of 2015, a first draft of the localization of the full human proteome became available for use with the continual addition of new protein expression data.
Tissue Microarrays
The TMA technology provides an automated array-based high-throughput technique in which as many as 1,000 paraffin-embedded tissue samples can be brought into one paraffin block in an array format. This allows for protein expression profiling on a large scale. Each antibody in the Human Protein Atlas project generates more than 500 high-resolution images corresponding to normal and cancer tissues. In this manner, an atlas for tissue expression and localization is built up for each protein with an available specific antibody. TMAs used within the Human Protein Atlas project include samples from 48 different human normal tissue types and 20 different types of cancer. Normal tissues are sampled from 144 different individuals and cancer tissues are derived from 216 unique tumors.1,2
TMAs are constructed by extracting cylinders of formalin fixed, paraffin embedded tissue from donor blocks with a sharp punch and assembling them into a recipient block with properly sized holes in a grid pattern (Figure 1). From one array block, ~250 sections can be achieved and prepared for IHC analysis.

Figure 1.Cylinders from donor blocks are extracted and inserted into a recipient block. A Donor blocks of formalin fixed, paraffin embedded human tissues. B Recipient block (Tissue Microarray) representing 48 different human normal tissue types ready to be sectioned and used for IHC analysis.

Figure 2.Schematic figure of the immunohistochemical staining reaction. Prestige Antibodies are used as primary antibodies and the secondary antibody is labeled with the enzyme HRP. HRP forms a complex with the substrate H2O2 and in the presence of the chromogen DAB, a brown color can be visualized using light microscopy. The signal can be amplified using an enzyme-linked dextran polymer (figure to the right).
Trained professionals determine the optimal dilution and approve antibodies based on a comparison of staining pattern, available information from gene and protein public databases, as well as in-house technical validation such as protein arrays and Western blots.
Scanning and Annotation
All immunostained slides are scanned to generate high-quality images. More than 7 terabyte of image data is generated each month. The images representing immunostained tissue sections (TMAs) are analyzed and annotated by certified pathologists using a web based annotation software. All images and annotations are published and freely available at the Human Protein Atlas portal.
Subcellular analysis using immunofluorescence compared to IHC
Similarly, Immunofluorescence (IF)-based imaging, with its advantage of high resolution and sensitivity, remains an established golden standard for visualization of proteins at a subcellular level. In addition, immunofluorescence allows the use of several different antibodies tagged with different fluorophores simultaneously, which enables a more detailed analysis of subcellular localization patterns (Figure 3). Data on where proteins are localized within a cell provides important information as to what basic functions a protein may have as well as mapping other possible interacting proteins.

Figure 3.Immunofluorescent staining showing nuclear localization of the transcription factor, GBX2 (green) in human prostate cancer (PC-3) cells using Prestige Antibodies® Anti-GBX2 antibody that was produced in rabbit.
Numerous studies based on immunofluorescence are often performed on cultured cells with the disadvantage of not being able to analyze cells in their natural tissue context. In addition to the subcellular protein profiling available through immunofluorescence, localization information at a subcellular level can also be achieved using Prestige Antibodies® in IHC (Figure 4). Figures 4 A-C show examples of immunohistochemical staining using Prestige Antibodies® for recognition of cell membrane-related proteins, Figures 4 D-F show examples of proteins expressed in different cytoplasmic compartments and Figures 4 G-I show proteins expressed in different nuclear structures.
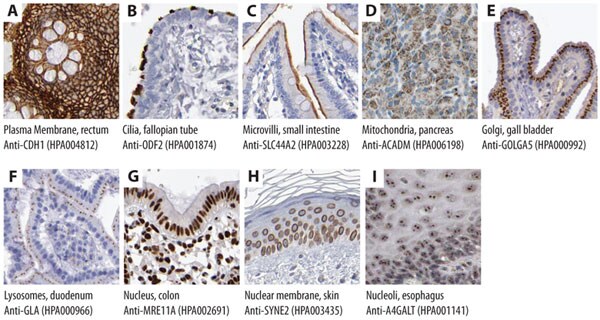
Figure 4.Immunohistochemical stainings showing subcellular localization using Prestige Antibodies®. Recognition of target antigen is represented by brown color.
Summary
- The use of Prestige Antibodies® in Immunohistochemistry on Tissue Microarrays (TMAs) has allowed for protein expression profiling in a large-scale format.
- In the Human Protein Atlas Project, TMAs including samples from 48 different human normal tissue types and 20 different types of cancer are used for protein localization analysis.
- For each Prestige Antibody®, more than 500 IHC images are publicly available on the Human Protein Atlas web portal
- By the use of Prestige Antibodies® in Immunohistochemistry studies, information on a subcellular level can be achieved.
About the Human Protein Atlas
The Human Protein Atlas is a public web portal managed by an academic project that aims to map the human proteome. More than 700 IHC, ICC, WB and IF images are presented for each antibody against human targets.
References
To continue reading please sign in or create an account.
Don't Have An Account?