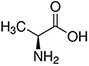
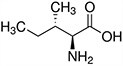
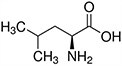
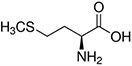
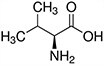
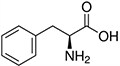
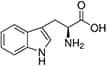
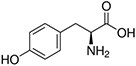
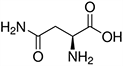
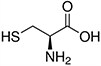
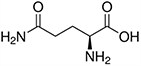
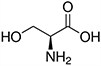
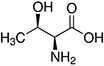
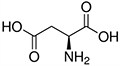
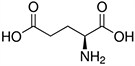
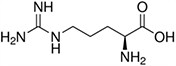
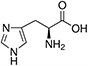
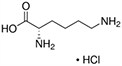
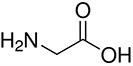
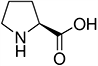
Amino acids are the compounds or building blocks that make up peptides and proteins. Each amino acid is structured from an amino group and a carboxyl group bound to a tetrahedral carbon. This carbon is designated as the α-carbon (alpha-carbon).
Amino acids differ from each other with respect to their side chains, which are referred to as R groups. The R group for each of the amino acids will differ in structure, electrical charge, and polarity.
Refer to the charts and structures below to explore amino acid properties, types, applications, and availability.
Amino Acids with Hydrophobic Side Chain – Aliphatic
Amino Acids with Hydrophobic Side Chain – Aromatic
Amino Acids with Polar Neutral Side Chains
Amino Acids with Electrically Charged Side Chains – Acidic
Amino Acids with Electrically Charged Side Chains – Basic
Unique Amino Acids
1 pKa is the negative of the logarithm of the dissociation constant for the -COOH group.
2 pKb is the negative of the logarithm of the dissociation constant for the -NH3 group.
3 pKx is the negative of the logarithm of the dissociation constant for any other group in the molecule.
4 pl is the pH at the isoelectric point.
Reference: D.R. Lide, Handbook of Chemistry and Physics, 72nd Edition, CRC Press, Boca Raton, FL, 1991.
Hydrophobicity Index for Common Amino Acids
The hydrophobicity index is a measure of the relative hydrophobicity, or how soluble an amino acid is in water. In a protein, hydrophobic amino acids are likely to be found in the interior, whereas hydrophilic amino acids are likely to be in contact with the aqueous environment.
The values in the table below are normalized so that the most hydrophobic residue is given a value of 100 relative to glycine, which is considered neutral (0 value). The scales were extrapolated to residues which are more hydrophilic than glycine.
ApH 2 values: Normalized from Sereda et al., J. Chrom. 676: 139-153 (1994).
BpH 7 values: Monera et al., J. Protein Sci. 1: 319-329 (1995).
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?