MISSION® esiRNA Specifications
Custom & Predesigned esiRNA
Beginning in late October / early November 2020, esiRNA products that are delivered in tubes — Individual esiRNA, esiOPEN, and esiSEC — will begin shipping lyophilized instead of in solution (esiRNA products in plates — esiLibrary and esiFLEX — will continue to ship in solution). We are making this change to products in tubes (most esiRNA ships in tubes) for the following reasons:
- Lyophilized materials can be shipped at ambient temperature instead of frozen on dry ice. This reduction in shipping expenses will help us to keep product prices as low as possible.
- By removing dry ice from shipments, we will use less packaging, which in turn will help us meet our company’s commitment to reducing its environmental impact. As RNA, esiRNA is stable at room temperature for at least 3 weeks. Upon receiving your esiRNA, please place it in -20 °C storage until you are ready to use it. Appendix 2 in the product Technical Bulletin contains a resuspension protocol. There will be a transition period in which some esiRNA may still ship in solution at the same time as some ships dry. We will expedite the shift to all units in dry form as quickly as possible.
All ordering and quotation options are in the esiRNA Specifications table
MISSION® esiRNA—powered by Eupheria Biotech—provide RNAi researchers with a proven, cost-effective, and simple way to perform post-transcriptional silencing of protein-coding genes and lncRNA (long non-coding RNA). Biologically-prepared, esiRNA are comprised of a heterogeneous pool of siRNA (natural RNA, no modifications) that all target the same mRNA sequence. These multiple silencing triggers lead to highly specific and effective gene knockdowns with lower off-target effects than single, chemically-synthesized siRNA (Figure 1).

Figure 1.Knockdown of target mRNA can be accomplished by chemically-synthesized siRNA or enzymatically-prepared siRNA (esiRNA). A) Chemically-synthesized siRNA consists of a single silencing trigger of 21 bp that is complementary to the target mRNA. The high concentration of the siRNA in the transfection reaction leads to pronounced off-target effects. B) In contrast, esiRNA consists of a pool of hundreds of siRNA (21 bp) that cover a region of 300 – 600 bp of the target mRNA. Each individual siRNA has a lower concentration in the pool, leading to lower off-target effects, while producing an efficient knockdown.
Product Benefits
- Guaranteed gene silencing
- Lower off-target effects than single, chemically-synthesized siRNA
- High on-target specificity makes for an effective primary screening tool (Figure 2)
- Affordable genome-scale RNAi screening tool
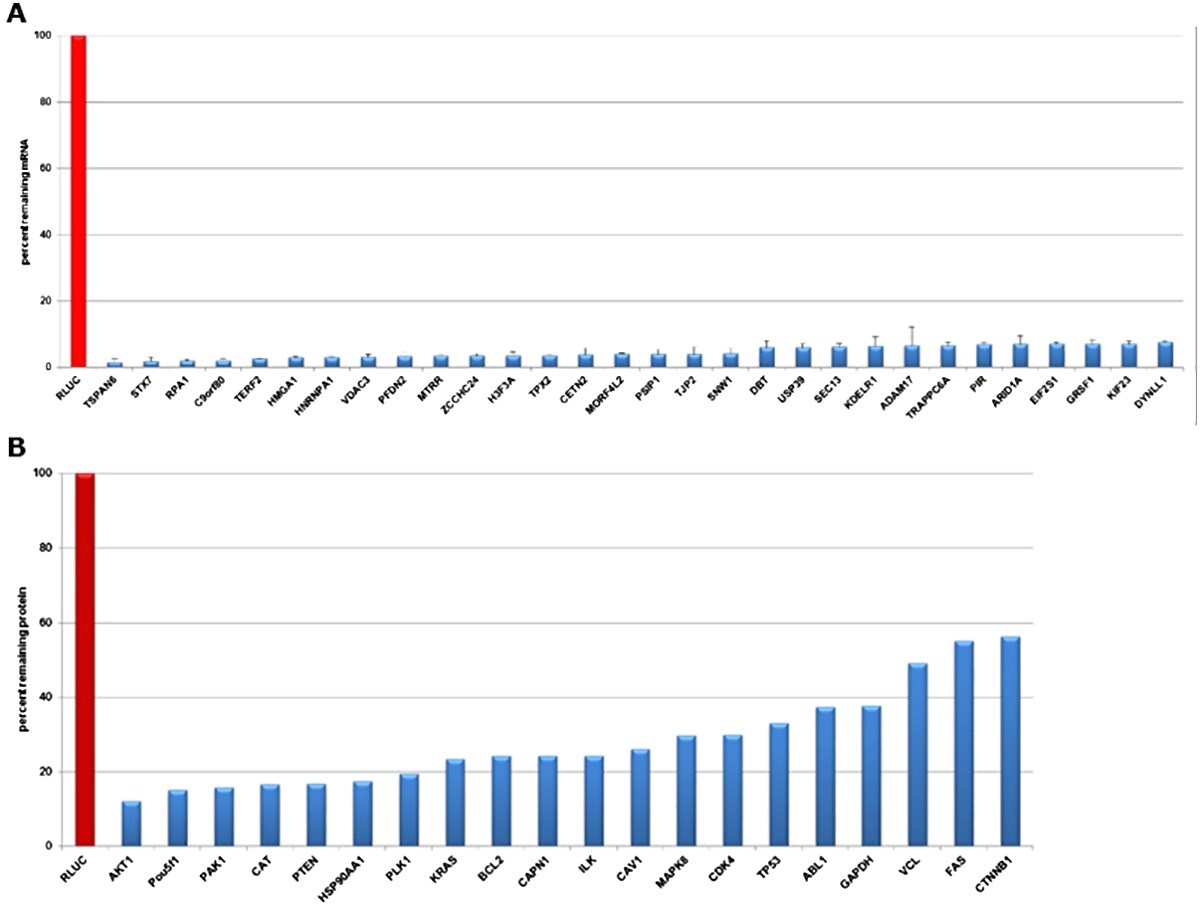
Figure 2.Evaluation of gene knockdown in HeLa cells transfected with esiRNA directed against Renilla Luciferase (negative control) and different expressed targets at mRNA and protein levels. A) qPCR data validate knockdowns 24 hours post-transfection. The mRNA abundance was reduced by >90% in all transfections. B) Quantitative Western Blot data validate efficient knockdown 72 hours post-transfection. Protein levels were reduced 45 to 90%.
Product Features
The following apply to all product options presented in the esiRNA specifications table:
- Purification: Q-Sepharose separation, isopropanol precipitation & ethanol washing
- Sequence Form: Pool of hundreds of siRNA with an average duplex length of 21 bp
- Quality Control: Conducted at two stages
o PCR product of cDNA clone is analyzed by gel electrophoresis & DNA sequencing
o Digestion reaction is analyzed by gel electrophoresis - Stability: Viable for 2 years when stored at -20 °C
For anything marked as ‘Inquire’ below or If you have needs that are different from the other general specifications presented, please send a request to sirnarequest@milliporesigma.com.
Browse the complete list of predesigned esiRNA.
Control esiRNA
A variety of positive and negative controls are available (Figure 3):
- Positive controls: Human Eg5 (KIF11) & mouse Eg5 (Kif11) esiRNA as well as all validated esiRNA (see table below) to ensure transfection & experimental set up optimization
- Negative controls: RLUC, FLUC & EGFP (EGFP can also be used as a positive control in cells expressing EGFP) esiRNA to distinguish sequence-specific silencing from non–specific effects

Figure 3.Phenotypic analysis of HeLa cells transfected with esiRNA against A) RLUC (negative control) and B) Eg5/KIF11 (positive control). RLUC does not induce any phenotypic changes, while Eg5/KIF11 induces mitotic arrest, shown by rounded cells.
Validated esiRNA
Many common gene targets have been validated for ≥70% mRNA knockdown (qPCR and Western Blot validation data). See the table for a list of commonly-ordered, validated esiRNA by gene symbol. Validated esiRNA are suitable for transfection optimization and as positive controls.
lncRNA esiRNA
Transcriptome analyses reveal that up to 90% of the genome is transcribed into non-coding RNA, which among other types, includes lncRNA. The lncRNA are associated with different functions, including chromatin modification, co-activation of transcription factors, transcription, interaction with RNA-binding proteins, and repression of promoters. In addition, lncRNA have been implicated in diseases such as cancer. The details of how lncRNA work must be further elucidated. The esiRNA are an effective screening tool to study lncRNA function (Figure 4).

Figure 4.qPCR data validating knockdown of lncRNA in HeLa cells 24 hours post-transfection with corresponding esiRNA. Controls were transfected with esiRNA against RLUC and used for normalization.
Product Manufacturing
Biologically-prepared (Figure 5), esiRNA is manufactured by cleaving long dsRNA (double-stranded RNA) with the endoribonuclease, E. coli RNase III

Figure 5.Manufacturing overview of esiRNA for single genes. A) DEQOR is used to select the mRNA target region that will produce the largest number of highly-effective siRNA, cover all known transcript variants, and minimize off-target effects. B) The target region is extracted from the cDNA clone and amplified via PCR (both primers are used to introduce T7 promoters. C) The PCR product is transcribed in vitro utilizing RNA polymerases. D) The annealed and long, double-stranded RNA (dsRNA) is digested with RNase III, and purification removes the DNA template, residual NTPs, and incompletely digested dsRNA. Finally, esiRNA is ready as a pool of hundreds of siRNA with an average duplex length of 21 bp.
esiRNA Guarantee
MISSION® esiRNA will reduce target mRNA levels by 70%, in cultured cells, when transfected at a concentration of ≥30 nM. If the esiRNA does not knock down the target gene by 70%, we will provide an additional esiRNA for that gene, free of charge. If there are no more esiRNA for that gene, we will refund the purchase price.
Receipt of appropriate supporting data for transfection efficiency is required for the guarantee. Appropriate supporting data for transfection efficiency would include qPCR data comparing target mRNA levels of a validated MISSION® esiRNA, transfected at ≥30 nM, to an appropriate negative control (such as mock transfection, RLUC, FLUC, or eGFP esiRNA), demonstrating knockdown of the target mRNA of 70%.
Due to the variability of antibodies and protein half-lives, we are unable to accept data from protein-based detection methods.
Frequently Asked Questions
To learn more about esiRNA, visit our Frequently Asked Questions section.
Video Tutorial
Learn about screening in Mammalian cells with esiRNA.
If additional help is needed, please consult our technical services group at oligotechserv@merckgroup.com.
Learn more about RNA interference: from single gene studies to whole-genome screens - Dr. Julia Krüger Senior Scientist at Eupheria Biotech GmbH
If additional help is needed, please consult our technical services group at oligotechserv@sial.com.
Select Citations
Single-Gene esiRNA
Whole Genome & Sub-Library Screens
lncRNA esiRNA
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?