Successful Amplification of Thermally Degraded DNA Using PCRboost™
Product Nos. 63301-001, 63301-011, 62301-021
Introduction
DNA samples are often exposed to harsh environmental conditions, including heat, humidity, UV or chemicals. Some procedures, such as the extraction of samples from formalin-fixed paraffin embedded tissue samples, subject DNA to multiple extreme conditions. The common end result of exposure to these varied conditions is severe structural degradation and random fragmentation of the DNA. Degraded DNA poses a problem for various necessary analytical methods such as PCR amplification of larger fragments, because intact fragments spanning the primer region are underrepresented in the population of fragments. PCRboost™ was designed to enhance the amplification of such DNA without the need for time consuming optimizations as would normally be the case. PCRboost successfully amplifies thermally degraded DNA resulting in a visible amplicon that can be used for downstream applications such as cloning and sequencing.
Materials and Methods
Artificial DNA degradation: Genomic DNA (gDNA) was purified from human blood collected in EDTA tubes and purified using the QIAamp DNA Blood Mini Kit. 500ng gDNA were exposed to 70 °C for 20h and the degradation pattern visualized by agarose gel electrophoresis (0.8%).
PCR amplification: 1ul (~50ng) of the thermally degraded DNA or control DNA stored at 4 C were used as template for PCR reactions. Reactions were set up using 2.5 U Taq DNA polymerase (NEB), 3µL 10x thermopol reaction buffer (NEB), 0.5 µL dNTPs (10µM each nucleotide), 0.5 µL each human ß-actin forward (5’ctacctcatgaagatcctcacc3’) and ß-actin reverse (5’gtacttgcgctcaggaggagc3’;10µM each) in a final volume of 30 µL. The volume in the thermally degraded DNA PCR reaction tubes was brought up either in water or in PCRboost. Cycling parameters were: 94 °C for 5 min followed by 40 cycles of 94 °C for 15 sec, 55 °C for 30 sec and 72 °C for 30 sec. 10 µL of each PCR reaction was run on a 0.8% agarose/ethidium bromide gel.
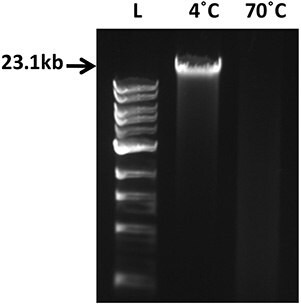
Figure 1.500ng human genomic DNA stored at 4 °C or thermally degraded at 70 °C for 20h was run on a 0.8% agarose. L: 1kb ladder (NEB).

Figure 2.50ng of cold-stored (4 °C) or thermally degraded DNA were used as template in PCR reactions amplifying the human ß-actin gene. PCR reactions were brought up to volume either in water or PCRboost. L: ladder.
Results and Discussion
PCRboost is a novel amplification enhancer. Here we tested the effects of PCRboost in amplification of severely degraded DNA samples. The addition of PCRboost resulted in a visible amplicon as visualized by agarose gel electrophoresis. The addition of PCRboost to PCR reactions has resulted in significant increases in amplicon yields, successful amplification of trace amounts of DNA or difficult to amplify DNA regions (eg GC-rich regions). The results presented in this paper suggest that PCRboost adds a great benefit to the analysis of degraded DNA.
To continue reading please sign in or create an account.
Don't Have An Account?