E1510
Ethidium bromide solution
BioReagent, for molecular biology, 10 mg/mL in H2O
Synonym(s):
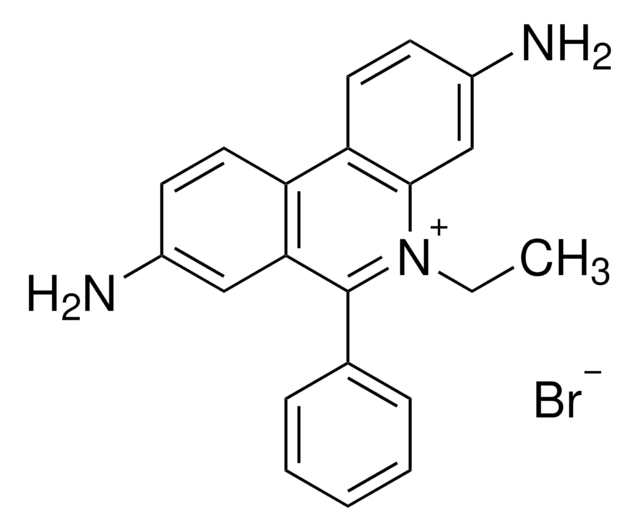
3,8-Diamino-5-ethyl-6-phenylphenanthridinium bromide, EtBr, Homidium bromide
About This Item
Recommended Products
grade
for molecular biology
Quality Level
product line
BioReagent
concentration
10 mg/mL in H2O
technique(s)
electrophoresis: suitable
suitability
suitable for gel electrophoresis
SMILES string
[Br-].CC[n+]1c(-c2ccccc2)c3cc(N)ccc3c4ccc(N)cc14
InChI
1S/C21H19N3.BrH/c1-2-24-20-13-16(23)9-11-18(20)17-10-8-15(22)12-19(17)21(24)14-6-4-3-5-7-14;/h3-13,23H,2,22H2,1H3;1H
InChI key
ZMMJGEGLRURXTF-UHFFFAOYSA-N
Looking for similar products? Visit Product Comparison Guide
Application
- as a stain to visualize U937 cells to assess cell viability
- to detect polymerase chain reaction products
- in agarose gel electrophoresis based gel retardation assay
Biochem/physiol Actions
Reconstitution
related product
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Acute Tox. 3 Inhalation - Muta. 2
Storage Class Code
6.1D - Non-combustible acute toxic Cat.3 / toxic hazardous materials or hazardous materials causing chronic effects
WGK
nwg
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Protocols
The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) system was discovered in bacteria, where it functions as an adaptive immune system against invading viral and plasmid DNA.
The GenElute Mammalian Genomic DNA Purification Kit Protocol describes the isolation of pure, high molecular weight DNA from a variety of mammalian sources.
GenElute Bacterial Genomic DNA Kit protocol describes a simple and convenient way for the isolation of pure genomic DNA from bacteria.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service