Protein Binding Determination - Comparison Study of Techniques & Devices
M. James Ross, Senior R&D Scientist,
Olga Shimelis, R&D, R&D Manager Sample Preparation,
Candace Price, Product Manager Sample Prep – SPE and BioSPME
Introduction

An important aspect of drug discovery is understanding the interaction of the drug candidate with plasma proteins and lipids. The binding of drugs to the proteins and lipids is referred to as the plasma protein binding (PPB or Fb). The molecular attributes of a drug can provide useful insights into its number of interactions with the protein. In general, organic acids have a single binding site with albumin, whereas organic bases have multiple bindings sites associated with glycoproteins.1 In addition to albumin, other proteins commonly associated with drug binding are alpha-1-acid glycoprotein (AAG) and lipoproteins, such as very high-density lipoprotein (VHDL) and low-density lipoprotein (LDL).2 When measuring the pharmacologic potency of a drug, it is the free fraction or unbound fraction (FU) of a drug that is generally considered responsible for its activity as described by the free drug hypothesis.3,4,5
Determining the protein binding properties of a drug is important to understand the amount of free drug available in the blood. So far, equilibrium membrane dialysis has been the traditional technique used to measure drug protein binding. The technique involves equilibration of a drug rich plasma sample with a drug- free buffer across a membrane. This allows the free drug to migrate across the membrane and prevents the protein bound drug from moving into the buffer. This equilibrium takes more than 24 hours to establish. Other techniques such as rapid equilibrium dialysis further reduce the workflow time from >24 hours to 6 hours by using specifically designed devices.
In this study, the Supel™ BioSPME 96-Pin device is used to measure drug protein binding. Supel™ BioSPME 96-Pin devices have been developed using solid phase microextraction, SPME, technology to extract free unbound analytes from biological fluids. These devices consist of a 96-pin plate, with the tips of pins coated with a thin layer of adsorbent particles. The patented binder within the coating allows the small analytes of interest to bind, while preventing macromolecules from binding. This enables a robust, selective, and non-exhaustive extraction of free analytes, having both qualitative and quantitative applications. The 96-pin configuration allows direct sampling from 96-well plates and is compatible with robotic liquid handling systems, providing a fully automated high-throughput methodology.
In this study, the Supel™ BioSPME 96-Pin device workflow is compared with a rapid equilibrium dialysis technique to measure drug protein binding. A series of compounds with molecular weights in the range of 230-750 Da and a hydrophobicity (LogP) range of 1.5-5 were utilized for comparison of protein binding values and determined using each technique. In addition to measured values and accuracy, the study compared overall sample cleanliness and workflow time of each technique.
Experimental Method
Determination of Protein Binding using Supel™ BioSPME 96-Pin Device
Human plasma and phosphate buffered saline (PBS) were spiked at a therapeutically relevant concentration and incubated for one hour at 37 ºC while shaking at 300 rpm. After the incubation, 200 μL of plasma and PBS were loaded into wells in separate columns of an extraction well plate (n = 8). The protein binding determination workflow using the Supel™ BioSPME 96-Pin device was conducted with an automated robotic liquid handling system. Briefly described in Figure 1, the pin device is statically (without shaking) conditioned for twenty minutes in isopropanol, then transferred into a new well plate containing water for 10 seconds (wash step). This is followed by the extraction step, where the pin device is transferred into the preloaded extraction well plate and the analyte extraction takes place while shaking at 1200- 1250 rpm at 37 ºC for 15 minutes. The pin device is returned to the water solution for a 60 second wash and then transferred into a desorption well plate for the final step. The desorption solution is a mixture of 80:20 methanol:water, and the pin device undergoes desorption for 20 minutes under static conditions.
Figure 2 shows how a Supel™ BioSPME 96-Pin device being maneuvered by the automated liquid handing system gripper.
Figure 2. Supel™ BioSPME 96-pin device on automated liquid handing system.
Figure 3 represents the extraction of free unbound analyte onto the Supel™ BioSPME 96-Pin device. The amount extracted does not greatly impact the concentration of free analyte and is termed non- depletive. As the buffer solution is considered 100% free, the Supel™ BioSPME 96-Pin device extracts more from buffer than from plasma.
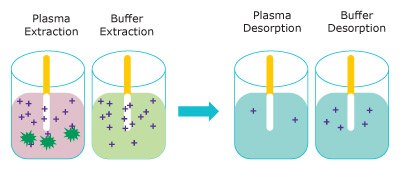
Figure 3. Representation of the extraction step (left) removing free analytes from plasma (pink) and buffer (blue) and the analytes releasing into the desorption solution (right). (dark green = proteins, purple = analyte)
The extraction plates used in this study included both plastic and glass-coated. The choice of the plate depended on the properties of the compound and how well the compound behaved in buffer solution. More hydrophobic compounds, such as ketoconazole and imipramine were found to exhibit non-specific binding to plastic and thus had better extraction efficiency from glass-coated 96-well plates. Extractions of erythromycin and propranolol were also performed using glass-coated plates because of their higher extraction efficiency values, in comparison to extraction from plastic plates.
Protein Binding Determination by Rapid Equilibrium Dialysis
Rapid equilibrium dialysis was performed as directed by the product instruction sheet. Two hundred microliters of human plasma “spiked” at a therapeutically relevant concentration and 400 μL of phosphate buffered saline (PBS) were loaded into corresponding chambers of the rapid equilibrium dialysis device, in at least triplicate sets. Dialysis proceeded for at least 4 hours while covered and shaking at 300 rpm and 37 ºC on an Eppendorf® shaker. At the end of dialysis, 50 μL of the spiked plasma was mixed with 50 μL of clean (unspiked) PBS, and 50 μL of the dialysate (buffer compartment) was mixed with 50 μL of clean plasma. This was done to ensure matrix consistency. Next, protein precipitation was initiated with the addition of 300 μL of ice-cold acetonitrile to each sample before centrifugation at 5,000 rpm for 10 minutes at 4 ºC. Finally, the supernatant was transferred into glass vials for analysis by LC-MS/MS.
Samples prepared using both the Supel™ BioSPME 96-Pin device and the rapid equilibrium dialysis device were analyzed by LC-MS/MS. The chromatographic and mass spectrometric analyses were performed on an Agilent 1290 / AB Sciex 6500 LC-MS/MS system following the conditions described in Table 1. Quantitation was performed using an external calibration in the desorption solution.
Results & Discussion
The Supel™ BioSPME method determines the free concentration of analyte in plasma by comparing it with the extraction of the same analyte from buffer samples, where 100% of the analyte is considered to be free of protein binding. Supel™ BioSPME 96-pin devices were directly compared with a rapid equilibrium dialysis technique, as it is often considered the standard approach and workflow of choice for plasma protein binding determination. Supel™ BioSPME 96-pin devices show numerous advantages over the rapid equilibrium dialysis technique, in terms of time savings, sample cleanliness, and a simplified workflow, while still maintaining the same high standards of accuracy and reproducibility needed by the bioanalytical laboratories performing this analysis. A comparison of the protein binding values obtained using the rapid equilibrium dialysis method and Supel™ BioSPME method is shown below in Figure 4.
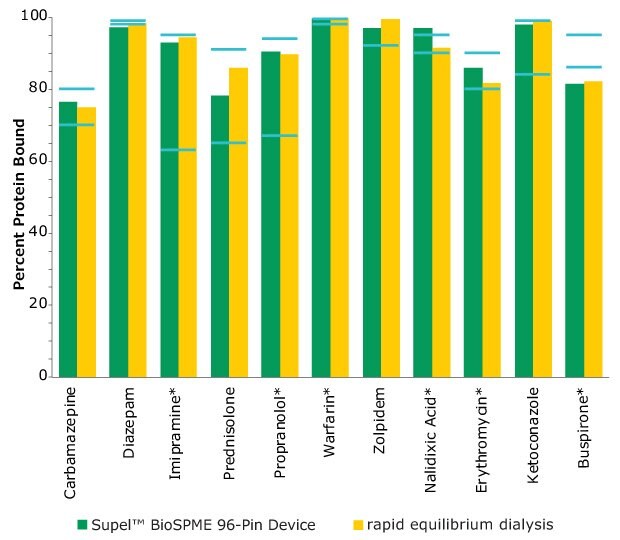
Figure 4. Comparison of Protein Binding Values Between Supel™ BioSPME 96-Pin Device and Rapid Equilibrium Dialysis Methods. The blue lines indicate the published protein binding literature value intervals. Compounds with asterisks are charged at physiological pH.
The data from Figure 4 is shown in tabular form in Table 2. The values from the BioSPME method are in good agreement with values determined using rapid equilibrium dialysis devices and the reported literature values.
Comparison of Workflow Time: Supel™ BioSPME Device vs Rapid Equilibrium Dialysis
As high throughput laboratories are always interested in optimizing efficiencies as much as possible, the time to perform each one of the workflows was evaluated for comparison purposes. The Supel™ BioSPME 96-Pin device workflow (<2 hours) takes one third of the amount of time the rapid equilibrium dialysis workflow (6 hours) takes, as shown in Table 3. This provides the opportunity to increase throughput three-fold, allowing the liquid handling instrument to be used for other assays, and giving the scientist time for other projects.
Conclusion
The Supel™ BioSPME 96-Pin device technique has shown significant time savings for protein binding determination when compared with the rapid equilibrium dialysis method. Since the workflow for the Supel™ BioSPME 96-Pin device is less than two hours compared to the rapid equilibrium dialysis method, it triples the throughput. In addition, the format of the Supel™ BioSPME 96-Pin device allows for a fully automated robotic method, without the need for additional hardware, i.e. a centrifuge, as required when performing the rapid equilibrium dialysis method. This translates to increased productivity and reduced manual steps for the laboratory. The accuracy of the protein binding values obtained using the Supel™ BioSPME 96-Pin device are in agreement with those obtained using the rapid equilibrium dialysis method; as demonstrated with 10 compounds with varying LogP values. The patented binder and coating of the Supel™ BioSPME 96-Pin device allows for selective extraction of target analytes, while excluding larger macromolecules, to provide a fast and accurate drug protein binding measurement.
See also the full application note at SigmaAldrich.com/BioSPME
References
To continue reading please sign in or create an account.
Don't Have An Account?