Quantification of Host Cell Proteins in Biotherapeutics using Stable Isotope Labeled Chinese Hamster Ovary Proteins (SIL-CHOP) as an Internal Standard
Residual presence of host cell proteins (HCPs) in recombinant therapeutic products has considerable clinical safety risks associated with a potential immunological response in patients. Although traditional methods, such as anti-HCP-ELISA, are well accepted for quantification, these methods may underestimate the amount of HCPs. Not all HCPs provoke the same immune responses during antibody generation. Further, interaction of low abundance HCPs with high abundance therapeutics may limit detection of HCPs with ELISA’s antibodies. To enable a more comprehensive characterization and quantification of HCPs, we have developed a SIL-CHOP (stable isotope labeled Chinese hamster ovary proteins) internal standard spanning a wide dynamic range of abundance and demonstrate its utility in identification and quantification of HCPs in downstream process development samples via LC-MS.
Methods
CHO-K1 cells were cultured in serum-free medium enriched with 13C6 15N4-Arg and 13C6 15N2-Lys. Proteins from the host cell culture fluid were isolated and the total protein concentration was determined using BCA assay. For characterization of SIL-CHOP, the sample was reduced, alkylated and digested with trypsin. The digested peptides were fractionated in a spin column format using high-pH reversed-phase SPE. Eight resulting fractions were analyzed using low pH C18-nano-LC-MS/MS. The isotopic incorporation was calculated to be greater than 98%. The data was searched against the Uniprot Cricetulus Griseus database and relative abundance of each protein was calculated based on ion intensities. The SIL-CHOP was spiked into downstream process development samples and SigmaMAb antibody standard and the amount of each HCP was determined in each sample.

Figure 1.HCP Characterization and Quantification Workflow
Characterization of SIL-CHOP

Figure 2.Incorporation of SIL for two peptides from CHO proteins.
2370 Unique SIL-CHO Proteins Identified

Figure 3.The Gene Ontology biological process terms of identified SIL-CHO proteins.

Figure 3.Identified SIL-CHO proteins arranged by relative abundance based on total ion intensity. HCPs identified in published analyses are highlighted.1-3
Detection of HCP Using SIL-CHOP Internal Standard

Figure 4.Protein IDs from four downstream purification samples for SIL-CHOP and unlabeled HCPs. Variation in the number of SIL-CHO proteins identified indicates a matrix effect across the four samples, impacting the ability uniformly to detect unlabeled HCP proteins.
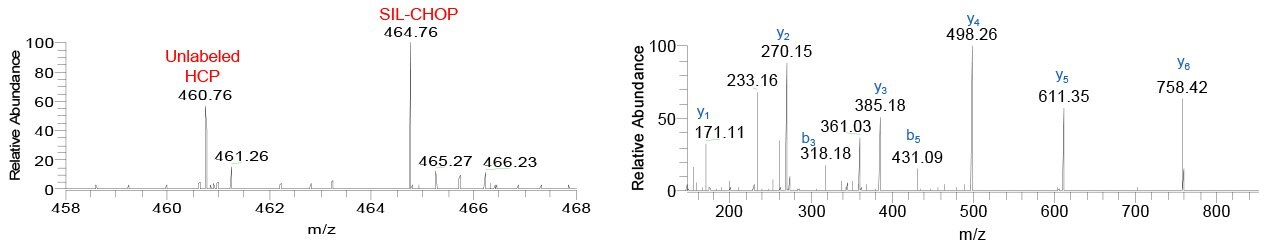
Figure 5.MS spectrum showing unlabeled GLFIIDDK (m/z 460.76) and SIL-GLFIIDDK (m/z 464.76) derived from Peroxiredoxin-1, PRDX1, in the AEX-purified sample spiked with SIL-CHOP, and corresponding MS/MS spectrum. This peptide was not identified by database search, but was manually identified by presence of the heavy/light peak pair and MS/MS match to SIL-CHOP spectral library.
Quantification of HCP Using SIL-CHOP Internal Standard

Figure 6.SIL-CHOP internal standard was spiked into SigmaMAb antibody standard (MSQC4) at 100, 1000, and 10,000 ppm (ng/mg), corresponding to SIL-Histone H3 spike levels of 0.43 to 43 ppm based on label-free quantitation of SIL-CHOP standard. The amount of Histone H3 in SigmaMAb was calculated to be ~10 ppm based on XIC peak area ratio of the labeled and unlabeled peptides, in blue and red, respectively.
Summary
- An SIL-CHOP standard has been prepared with broad coverage and isotopic incorporation >98%.
- The use of SIL-CHOP as an internal standard in quantitative mass spectrometric based assays of HCPs has been demonstrated.
Um weiterzulesen, melden Sie sich bitte an oder erstellen ein Konto.
Sie haben kein Konto?