Restriction Enzymes
Introduction
Restriction endonucleases popularly referred to as restriction enzymes, are ubiquitously present in prokaryotes. The function of restriction endonucleases is mainly protection against foreign genetic material especially against bacteriophage DNA. The other functions attributed to these enzymes are recombination and transposition. Restriction endonucleases make up the restriction-modification (R-M) systems comprised of endonuclease and methytransferase activities. The endonuclease recognizes and cleaves foreign DNA on the defined recognition sites. The methyltransferase modifies the recognition sites in the host DNA and protects it against the activity of endonucleases. The sequences in foreign DNA are generally not methylated and are subjected to restriction digestion. Each restriction enzyme recognizes a specific sequence of 4–8 nucleotides in DNA and cleaves at these sites. Endonucleases isolated by different organisms with identical recognition sites are termed isoschizomers.
Nomenclature
Different bacterial species synthesize endonucleases depending on the infecting viral DNA. The guidelines for naming restriction enzymes are based on the original suggestion by Smith and Nathans.1 The enzyme names begin with an italicized three-letter acronym; the first letter of the acronym is the first letter of the genus of bacteria from which the enzyme was isolated, the next two letters are the two letters of the species. These are followed by extra letters or numbers to indicate the serotype or strain, a space, then a Roman numeral to indicate the chronology of identification. For example, the first endonuclease isolated from Escherichia coli, strain RY13 is named as EcoR I. Hind III is the third endonuclease of four isolated from Haemophilus influenza, serotype d.
Factors that affect Restriction Enzyme Activity
The digestion activity of restriction enzymes depends on the following factors:
- Temperature: Most endonucleases digest the target DNA at 37 °C with few exceptions. Some work at lower temperatures (~25 °C, Sma 1) while Taq I works at 65 °C.
- Cofactors: Restriction endonucleases require certain cofactors or combination of cofactors to digest at the recognition site. All enzymes require Mg2+ as a cofactor for the endonuclease activity. In R-M systems with separate proteins having the restriction and methylation activities, S-adenosylmethionine (SAM) and ATP are required for methylation activity.
- Ionic Conditions: As mentioned previously, Mg2+ is required for all endonucleases but some enzymes also require ions such as Na+ and K+.
- Buffer systems: Most restriction enzymes are active in the pH range of 7.0–8.0. Tris-HCl, a temperature-dependent buffer, is the most commonly used buffer.
- Methylation status of DNA: Methylation of adenine or cytidine residues affects the digestion of DNA.
Classification of Restriction Enzymes
Restriction Enzymes are classified based on their activity sites, required cofactors, and recognition sequences. The detailed classification and description of each type of restriction endonucleases are presented in Table 1.
Products of Restriction Digestion
The digestion product of DNA strands may result in a fragment with either blunt ends or cohesive (sticky) ends at both the ends.
Cohesive 5' ends generated by EcoR I (Catalog Number R6265):

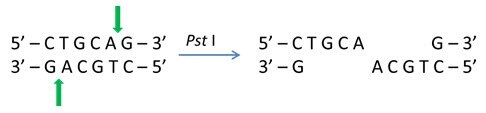
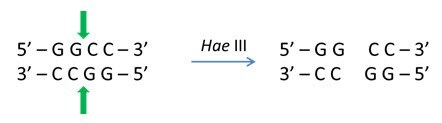
Methylation of DNA Strands
Methylation of DNA is a major factor that affects the specificity of restriction endonucleases. DNA isolated from strains of bacteria expressing methylases such as Dam or Dcm may be resistant to cleavage by endonucleases as the recognition sites are methylated. Plasmid DNA isolated from E. coli expressing Dam is methylated at the GATC sites making it resistant to cleavage by Mbo I. Certain methylation-specific endonucleases also degrade methylated DNA without affecting the methylated host DNA. BamH I isolated from Bacillus amyloliquefaciens H (Catalog Number R0260) cleaves methylated GGATCC sites of plasmid from E. coli expressing Dam. Dpn I (Catalog Number R8381) isolated from Diplococcus pneumonia targets methylated T7 DNA.
Star Activity of Restriction Enzymes
Star activity is defined as the alteration in the digestion specificity that occurs under sub-optimal enzyme conditions. Star activity results in cleavage of DNA at non-specific sites. Some of the sub-optimal conditions that result in star activity are as follows:
- pH >8.0
- glycerol concentration of >5%
- enzyme concentration >100 units/mg of DNA
- increased incubation time with the enzyme
- presence of organic solvents in the reaction mixture
- incorrect cofactor or buffer
Applications of Restriction Enzymes
Restriction endonucleases are widely used in molecular biology research for the following applications:
Genetic Engineering: The most popular application of restriction endonucleases is as a tool for genetic engineering. The endonuclease activity enables manipulation of the genome as well as introduction of sequences of interest in the host organism. This results in the production of the desired gene product by the host. This concept has wide range of applications in biotechnology in the production of antibiotics, antibodies, enzymes, and several secondary metabolites.
DNA mapping: DNA mapping using restriction enzymes (also known as restriction mapping) is a method to obtain structural information of the DNA fragment. In this technique the DNA is digested with a series of restriction enzymes to produce DNA fragments of various sizes. The resultant fragments are separated by agarose gel electrophoresis and the distance between the restriction enzyme sites can be estimated. This can be used to determine the structure of an unknown DNA fragment.
Gene Sequencing: A large DNA molecule is digested using restriction enzymes and the resulting fragments are processed through DNA sequencer to obtain the nucleotide sequence.
The other applications of restriction endonucleases include gene expression and mutation studies and examination of population polymorphisms.
References
Um weiterzulesen, melden Sie sich bitte an oder erstellen ein Konto.
Sie haben kein Konto?