miRNA (microRNA) Introduction
Introduction
Mature microRNAs (miRNAs) are a class of naturally occurring, small non-coding RNA molecules, about 21–25 nucleotides in length. MicroRNAs are partially complementary to one or more messenger RNA (mRNA) molecules, and their main function is to downregulate gene expression in a variety of manners, including translational repression, mRNA cleavage, and deadenylation. They were first described in 1993 by Lee and colleagues1, and the term microRNA was coined in 20012. Thousands of miRNAs have since been identified in various organisms through random cloning and sequencing or computational prediction. The miRBase, hosted by the Sanger Institute provides miRNA nomenclature, sequence data, annotation and target prediction information.
miRNA Biogenesis
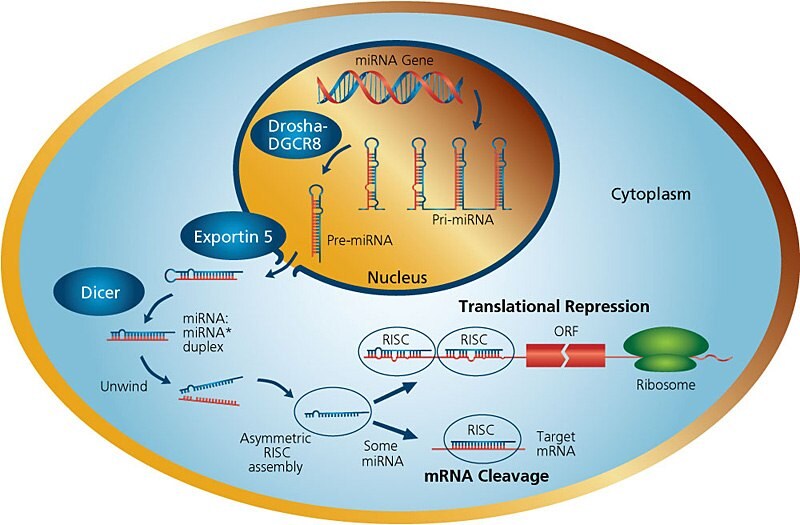
Figure 1.miRNA Pathways
The genes encoding miRNAs are much longer than the processed mature miRNA molecule. Many miRNAs are known to reside in introns of their pre-mRNA host genes and share their regulatory elements, primary transcript, and have a similar expression profile. For the remainder of miRNA genes that are transcribed from their own promoters, few primary transcripts have been fully identified. MicroRNAs are transcribed by RNA polymerase II as large RNA precursors called pri-miRNAs and comprise of a 5' cap and poly-A tail3. The pri-miRNAs are processed in the nucleus by the microprocessor complex, consisting of the RNase III enzyme Drosha4, and the double-stranded-RNA-binding protein, Pasha/DGCR85. The resulting pre-miRNAs are aproximately 70-nucleotides in length and are folded into imperfect stem-loop structures. The pre-miRNAs are then exported into the cytoplasm by the karyopherin exportin 5 (Exp5) and Ran-GTP complex6. Ran (ras-related nuclear protein) is a small GTP binding protein belonging to the RAS superfamily that is essential for the translocation of RNA and proteins through the nuclear pore complex7. The Ran GTPase binds Exp5 and forms a nuclear heterotrimer with pre-miRNAs6,8. Once in the cytoplasm, the pre-miRNAs undergo an additional processing step by the RNAse III enzyme Dicer9 generating the miRNA, a double-stranded RNA aproximately 22 nucleotides in length. Dicer also initiates the formation of the RNA-induced silencing complex (RISC)rev. in 10. RISC is responsible for the gene silencing observed due to miRNA expression and RNA interference11.
Dicer Function
Dicers are large 200 kDa proteins containing an ATPase/RNA helicase, a DUF283 (Domain of unknown function) domain, a PAZ (Piwi, Argonaut and Zwille) domain which can bind the characteristic two-base 3' overhangs of mi and siRNA, two catalytic RNase III domains (RIIIa and RIIIb), and a C-terminal double-stranded RNA-binding domain (dsRBD). Dicer functions as a monomer and has a single processing center with intramolecular dimerization of the two RNase III domains. Each RNase domain independently cuts one RNA strand of the duplex to generate products with 2-nt 3' overhangs. In addition to excising miRNAs from pre-miRNAs, Dicer enzymes process dsRNA to siRNAs. After Dicer cleavage, the miRNA pathway is similar to the central steps of RNA interference (RNAi) in animals. In contrast to siRNAs, microRNAs can direct RISC to down-regulate gene expression by translational repression (based on lower complementarity between miRNA and mRNA), or function as siRNAs do and mediate mRNA cleavage. The choice of post-transcriptional mechanisms is not determined by the origin of silencing RNA (siRNA or miRNA), but by the degree of complementarity. If the miRNA is perfectly or nearly complementary to its target, it can specifically cleave the target mRNA. Endogenously expressed miRNAs are usually imperfectly complementary to their target gene(s) and modulates the effect on gene expression via translational repression12.
RISC Complex
When Dicer cleaves the pre-miRNA stem-loop, two complementary short RNA molecules are formed, but only one is integrated into the RISC complex. RISC is a ribonucleoprotein complex containing members of the Argonaute (Ago) family of proteins. Argonaute proteins have endonuclease activity directed against mRNA strands that are complementary to their bound miRNA fragment. Argonaut is also partially responsible for selection of the guide strand and destruction of the passenger strand. The incorporated strand is known as the guide strand and is selected by the argonaute protein on the basis of the stability of the 5' end. The remaining strand, known as the passenger strand (*), is degraded as a RISC complex substrate. Selection of the guide strand from the dsRNA appears to be based primarily on the stability of the termini of the two ends of the dsRNA13. The strand with lower stability base pairing of the 2–4 nt at the 5' end of the duplex preferentially associates with RISC and thus becomes the active miRNA13. After integration into the active RISC complex, miRNAs exert their regulatory effects by binding to imperfect complementary sites within the 3' untranslated regions (UTRs) of their mRNA targets. The formation of the double-stranded RNA, resulting from the binding of the miRNA, leads to translational repression.
miRNA in Research
Although the first miRNA was identified over ten years ago, it is only recently that researchers have begun to understand the scope and diversity of these regulatory molecules. Growing evidence shows that miRNAs exhibit a variety of crucial regulatory functions related to cell growth, development, and differentiation, and are associated with a wide variety of human diseases. Several miRNAs have been linked to cancer14,15 and heart disease16. Expression analysis studies reveal perturbed miRNA expression in tumors compared to normal tissues15. MicroRNAs are deregulated in breast, lung, and colon cancer, and upregulated in Burkitt’s and other human B-cell lymphomas. As a consequence, human miRNAs are likely to be highly useful as biomarkers, especially for future cancer diagnostics, and are rapidly emerging as attractive targets for disease intervention. In addition to their link with cancer, microRNAs play an important role in the control of diverse aspects of cardiac function and dysfunction as well. Including myocyte growth, integrity of the ventricular wall, contractility, gene expression, and maintenance of cardiac rhythm. The misexpression of miRNAs has been shown to be necessary and sufficient for multiple forms of heart disease16.
miRNA Resarch Product Offerings
It is quite clear that miRNA research has great promise and may be the backbone of many of the most cutting-edge medical therapies of tomorrow. To keep up with the growing interest in miRNA, we have made a strong commitment to this rapidly developing area of research and is in the process of developing a comprehensive product portfolio to our customers including miRNA isolation, amplification, profiling, and functional analysis. The mirPremier microRNA Isolation Kit complements the already robust MISSION® RNAi product line which includes a broad choice of MISSION® siRNA, MISSION® miRNA mimics and shRNA products and services such as libraries, mRNA detection reagents, antibodies and AQUA™ peptides for protein level detection. An entire workflow of supporting reagents including cell culture, cell biology compounds and assays, transfection reagents, and much more).
miRNA Products
MISSION® Human miRNA Mimics
The Human miRNA Mimics library is based on MirBase ver. 21. Available in a library format (96-well plate format, 0.25 nmol/well) and in individual tubes (5 nmol). The novel MISSION® miRNA mimic design has been functionally tested for knockdown efficiency against natural miRNA targets and reduces possible off-target effects.
The MISSION® Target ID Library enables bench-top transcriptome-wide human miRNA and ncRNA gene target identification. With an innovative dual-positive selection system, it makes rapid whole transcriptome miRNA and ncRNA gene target screens available to any researcher with minimal time, reagent, or capital equipment expense.
MISSION® 3′UTR Lenti GoClone powered by SwitchGear Genomics™
We have partnered with SwitchGear Genomics™ to offer a genome-wide collection of MISSION® 3′UTR Lenti GoClones. The new system utilizes viral vectors to deliver human 3′UTRs fused to SwitchGear’s novel RenSP reporter gene. LightSwitch Luciferase Assay Reagent™'s have been designed specifically for use with RenSP and deliver maximum sensitivity, dynamic range, and convenience.
References
Um weiterzulesen, melden Sie sich bitte an oder erstellen ein Konto.
Sie haben kein Konto?