Purification from Unclarified Cell Lysate using HisTrap™ FF Crude
Extracted from Affinity Chromatography Vol. 2: Tagged Proteins, GE Healthcare, 2016
HisTrap™ FF crude is a ready-to-use column, prepacked with Ni Sepharose® 6 Fast Flow, for purification of histidine-tagged recombinant proteins. After thorough cell disruption, it is possible to load the unclarified, crude cell lysate directly on the column without centrifugation and filtration of the sample.
Direct loading of unclarified cell lysates decreases the total purification time and may increase the possibility of purifying sensitive target proteins without losing their activity.

Figure 3.14.HisTrap™ FF crude columns allow simple, one-step purification of histidine-tagged proteins without sample pretreatment such as centrifugation and filtration.
HisTrap™ FF crude columns are made of polypropylene that is biocompatible and does not interact with biomolecules. Columns are delivered with a stopper on the inlet and a snap-off end on the outlet. The columns have porous polyethylene top and bottom frits with a pore size optimized for loading of unclarified cell lysates directly on the column without causing back-pressure problems or leakage of the Ni Sepharose® 6 Fast Flow beads. Columns can be operated with a syringe and the supplied Luer connector, a peristaltic pump, or a chromatography system such as ÄKTA. Note that HisTrap™ FF crude columns cannot be opened or refilled.
See Figure 3.15 for a schematic of the protocol for HisTrap™ FF crude compared with conventional IMAC.
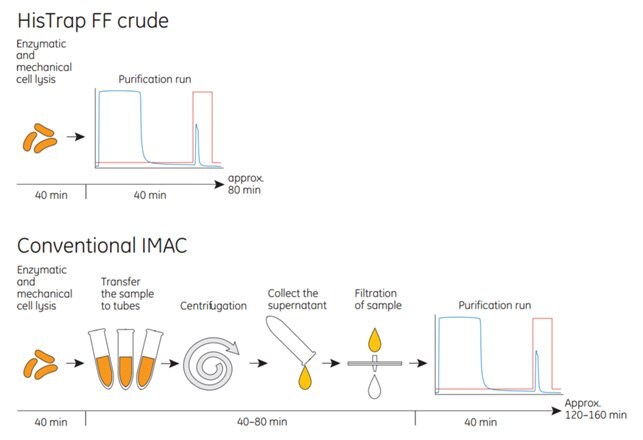
Figure 3.15.HisTrap™ FF crude columns save time during purification of histidine-tagged proteins compared with conventional IMAC.
Sample and buffer preparation
Refer to Purification using Ni Sepharose® 6 Fast Flow earlier in this chapter for a general procedure for sample and buffer preparation.
For direct loading of an unclarified sample, it is critical to obtain efficient cell lysis in order to avoid problems with back pressure. Apply the unclarified lysate on the column directly after preparation.
If the sample is too viscous, dilute it with binding buffer to prevent it from clogging the column; increase lysis treatment (sonication, homogenization); or add DNase/RNase to reduce the size of nucleic acid fragments.
If the sonicated or homogenized unclarified cell lysate is frozen before use, precipitation and aggregation may increase. New sonication of the lysate can then prevent increased back-pressure problems when loading on the column.
Purification
- Fill the pump tubing or syringe with distilled water. Remove the stopper and connect the column to the chromatography system tubing, syringe (use the provided Luer connector), or laboratory pump “drop to drop” to avoid introducing air into the system.
- Remove the snap-off end at the column outlet.
- Wash out the ethanol with 3 to 5 column volumes of distilled water.
- Equilibrate the column with at least 5 column volumes of binding buffer. Recommended flow rates are 1 mL/min or 5 mL/min for the 1 mL and 5 mL columns, respectively.
- Apply the unclarified lysate with a pump or syringe. Typical loading volumes of unclarified lysate (highly dependent on specific sample, sample pretreatment, and temperature at sample loading):
- HisTrap™ FF crude 1 mL: Up to 100 mL
- HisTrap™ FF crude 5 mL: Up to 500 mL
Note that the protein binding capacity may also limit the amount of sample that can be applied.
Continuous stirring of the sample during sample loading is recommended to prevent sedimentation. Sample loading at 4°C may increase the viscosity of the sample. An adverse effect of increased sample viscosity is that maximum back pressure for the column is reached at a lower sample volume loading on the column. Large volumes may increase back pressure, making the use of a syringe more difficult.
- Wash with binding buffer until the absorbance reaches a steady baseline (generally at least 10 to 15 column volumes).
- Elute with elution buffer using a step gradient or a linear gradient. For step elution, 5 column volumes of elution buffer is usually sufficient. A shallow gradient, for example, linear gradient over 20 column volumes or more, can separate proteins with similar binding strengths.
- After elution, regenerate the column by washing it with 5 to 10 column volumes of binding buffer. The column is now ready for a new purification of the same target protein.
The column does not need to be stripped and recharged between each purification if the same protein is going to be purified. Reuse of any purification column depends on the nature of the sample and should only be performed with identical tagged proteins to prevent cross-contamination. For more information on this topic and on cleaning and storage, refer to Appendix 1 (Characteristics of Ni Sepharose, Ni Sepharose® excel, TALON® Superflow, and uncharged IMAC Sepharose® products).
Unclarified lysates may cause increased air bubble formation during purification. An attached flow restrictor in the chromatography system can prevent this if it causes problems. If a flow restrictor is attached, it is important to change the pressure limit to 0.5 MPa (5 bar) on the ÄKTA system (where the column and the flow restrictor give a pressure of 0.3 MPa and 0.2 MPa, respectively).
Ni Sepharose® is compatible with reducing agents. However, we recommend removal of any weakly bound Ni2+ ions before applying buffer/sample that includes reducing agents. This can be accomplished by performing a blank run without reducing agents (see below). Do not leave or store HisTrap™ FF crude columns with buffers that include reducing agents.
Leakage of Ni2+ from Ni Sepharose® is low under all normal conditions. For very critical applications, leakage during purification can be even further diminished by performing a blank run (as described below) before loading sample.
Blank run:
Use binding buffer and elution buffer without reducing agents.
- Wash the column with 5 column volumes of distilled water (to remove the 20% ethanol).
- Wash with 5 column volumes of elution buffer.
- Equilibrate with 10 column volumes of binding buffer.
Application Examples
1. Purification of histidine-tagged protein expressed at low levels in Pichia pastoris using HisTrap™ FF crude
HisTrap™ FF crude columns are well-suited for purification of histidine-tagged proteins expressed at low levels from hosts such as Pichia pastoris. Using HisTrap™ FF crude columns, highly pure protein can be obtained directly from unclarified lysates of P. pastoris.
Figure 3.16 shows the purification of a histidine-tagged hydrolase from Saccharomyces cerevisiae expressed at low levels in P. pastoris. Unclarified sample was loaded directly onto a HisTrap™ FF crude 5 mL column without any centrifugation or filtration of the sample. Purity of the protein from the unclarified sample was high as determined by SDS-PAGE.

Figure 3.16.(A) c from Saccharomyces cerevisiae expressed in P. pastoris on HisTrap™ FF crude 5 mL. (B) SDS-PAGE under nonreducing conditions (ExcelGel SDS Gradient 8–18) shows the high purity obtained of the low-level expression protein.
2. Scale-up from 1 mL to 5 mL HisTrap™ FF crude
An experiment was performed to scale up from 1 mL to 5 mL HisTrap™ FF crude columns. The sample was unclarified E. coli extract containing MBP-(His)6, which had been prepared by enzymatic lysis in combination with homogenization prior to loading on the column. The samples contained approximately 8 and 40 mg of MBP-(His)6 for the 1 mL and 5 mL columns, respectively.
After chromatography (Figure 3.17A and 3.17B), SDS-PAGE analysis was performed. It shows (Figure 3.17C) that the purity and recovery (mg protein/mL medium) of the histidine-tagged protein purified on the two columns was almost identical.
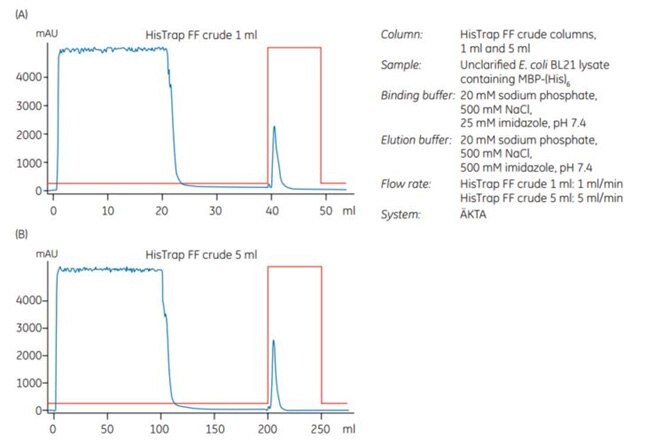
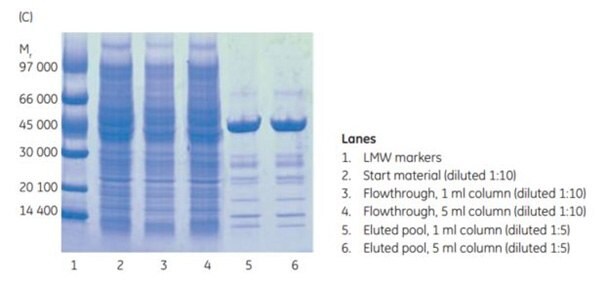
Figure 3.17.Scale-up from (A) 1 mL to (B) 5 mL HisTrap™ FF crude columns. Recovery of protein was 6.3 and 35.2 mg for the 1 mL and 5 mL columns, respectively. (C) SDS-PAGE under nonreducing conditions (ExcelGel SDS Gradient 8–18) confirms that scaling up from the 1 mL to the 5 mL column does not significantly affect the purification result.
3. Automated, two-step purification using HisTrap™ FF crude
HisTrap™ FF crude columns are well suited to run on ÄKTA systems such as ÄKTAxpress for high-throughput purification of histidine-tagged proteins. ÄKTAxpress enables automated, parallel purification of histidine-tagged proteins with the capacity to run a number of different multi-step protocols. A method wizard supplied with the UNICORN control software makes it easy to create methods for different purification protocols. Figure 3.18 shows an automated two-step purification of an unclarified lysate of E. coli containing MBP-(His)6. The first step in the purification protocol was AC, using HisTrap™ FF crude 1 mL column. The eluted peak from the affinity step was automatically collected in a loop and reinjected onto a HiLoad™ 16/60 Superdex 75 pg SEC column in the second step of the purification. Purity of the protein in fractions from the SEC step was confirmed by SDS-PAGE (Figure 3.18B).
The results show that HisTrap™ FF crude together with ÄKTAxpress facilitates and enables significant time savings in the purification of histidine-tagged proteins without compromising sample purity.
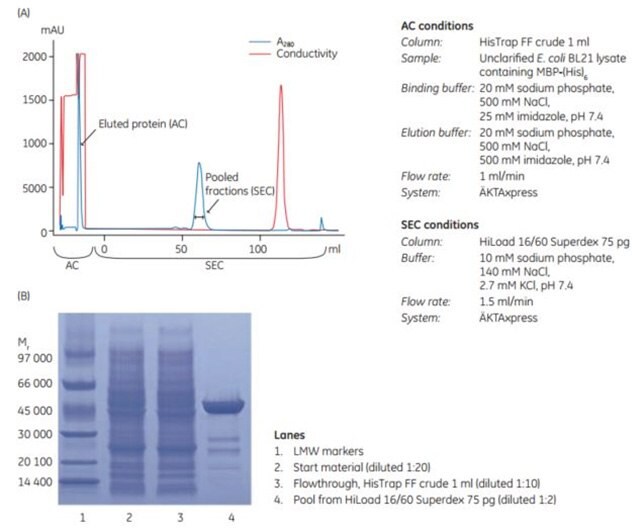
Figure 3.18.(A) Automated two-step purification of MBP-(His)6 from an unclarified lysate of E. coli using HisTrap™ FF crude and HiLoad 16/60 Superdex 75 pg on ÄKTAxpress. (B) SDS-PAGE was performed under nonreducing conditions on ExcelGel SDS Gradient 8–18.
To continue reading please sign in or create an account.
Don't Have An Account?