Prestige Antibodies® in Breast Cancer Research

- The Human Protein Atlas
- Prestige Antibodies® Powered by Atlas Antibodies
- Monoclonal Antibody Development
- Clinical Markers (ESR1, HER2, Ki67, PGR)
- Antibodies used in Breast Cancer Research
- Antibodies Against Gene Products in MammaPrint, Oncotype, EndoPredict and uPA Tests
- Antibodies Identified in the Human Protein Atlas

The Human Protein Atlas is Characterizing the Human Proteome
The Human Protein Atlas (HPA) project was initiated in 2003 by Swedish researchers, headed by Professor Mathias Uhlén, and funded by the Knut and Alice Wallenberg Foundation.1,2 It is a unique world leading effort performing systematic exploration of the human proteome using antibodies.
The aim of the HPA project is to present an expression map of the complete human proteome. To accomplish this, highly specific polyclonal antibodies are developed to all protein coding human genes and protein profiling is established in a multitude of tissues and cells using tissue arrays. Applications applied are immunohistochemistry (IHC), Western blot (WB) analysis, protein array assay and immunofluorescent based confocal microscopy (ICC-IF).
The Human Protein Atlas, November 2014
The 13th version of the Human Protein Atlas, released in November 2014, presents a tissue-based map of the complete human proteome. The extensive amount of data is divided into four separate 'sub atlases': the Tissue Atlas, the Cancer Atlas, the Subcell Atlas and the Cell Line Atlas. For all proteins represented in the Tissue Atlas, the expression profiles are based on IHC analysis on a large number of human tissues. The presentation of protein expression data in correlation to RNA sequencing data for each gene has now been included. In the Cancer Atlas, differentially expressed genes in several cancers can be studied, while the Subcell Atlas presents subcellular localization by confocal microscopy. Additional information about protein expression in common cell lines is included in the Cell Line Atlas, which has become an appreciated toolbox for research.
Tissue microarrays containing samples from 48 different normal human tissues, 20 different cancer types and 44 different human cell lines are utilized within the project. The 48 normal tissues are present in triplicate samples and represent 82 different cell types. All normal tissue images have undergone pathology-based annotation of expression levels and are displayed on the normal Tissue Atlas presenting information regarding the expression profiles of human genes both on mRNA and protein level. The mRNA expression data is derived from deep sequencing of RNA (RNA-Seq) from 27 major different normal tissue types.
The Cancer Atlas contains gene expression data based on protein expression patterns in a multitude of human cancer specimens. Altogether 216 different cancer samples, corresponding to the 20 most common forms of human cancer, have been analyzed for all included genes. All cancer tissue images have been manually annotated by pathologists and just as for the normal Tissue Atlas, protein data includes protein expression levels corresponding to 16.621 genes for which there are available antibodies.
Validation in Breast Tissue Samples and Cell Lines
IHC images from normal breast samples from three different individuals are available for each antibody in the normal Tissue Atlas. In addition, for each antibody, breast tumor samples from up to 12 patients in duplicates are presented in the Cancer Atlas and for the majority of the antibodies, also images from the MCF-7 and SK-BR-3 breast cell lines in the Cell Line Atlas.
References
Prestige Antibodies® Powered by Atlas Antibodies
Prestige Antibodies – the Building Blocks of HPA
The uniqueness and low cross reactivity of Prestige Polyclonals to other proteins are due to a thorough selection of antigen regions, affinity purification on the recombinant antigen, validation using several methods and a stringent approval process.
Development
The Prestige Antibodies are developed against recombinant human Protein Epitope Signature Tags (PrESTs) of approximately 50 to 150 amino acids. These protein fragments are designed, using a proprietary software, to contain unique epitopes present in the native protein suitable for triggering the generation of antibodies of high specificity. This is achieved by a complete human genome scanning to ensure that PrESTs with the lowest homology to other human proteins are used as antigens.
Approval
The approval of the Prestige Antibodies relies on a combined validation of the experimental results using IHC, WB or ICC-IF, from RNA sequencing and from information obtained via bioinformatics prediction methods and literature. Since the literature is often inconclusive, an important objective of the HPA project has been to generate paired antibodies with non-overlapping epitopes towards the same protein target, allowing the results and validation of one antibody to be used to validate the other one.
Prestige Antibodies Catalog
Today, there are more than 17,000 Prestige Polyclonals with 2,000 new antibodies added each year.
The antibodies developed and characterized within the Human Protein Atlas project are available to the scientific community as Prestige Antibodies and Atlas Antibodies.
Monoclonal Antibody Development
Prestige Antibodies also include a selected number of mouse monoclonal antibodies. The monoclonal catalog is regularly expanding with new products every year.
Unique Features
Special care is taken in offering clones recognizing only unique nonoverlapping epitopes and/or isotypes. Using the same stringent PrEST production process and characterization procedure as for the polyclonals, the monoclonal antibodies offer outstanding performance in approved applications, together with defined specificity, secured continuity and stable supply. In general they also permit high working dilutions and contribute to more standardized assay procedures.
Clone Selection
Functional characterization is performed on a large number of ELISA positive cell supernatants to select the optimal clones for each application prior to subcloning and expansion of selected hybridomas.
Epitope Mapping
Clones are epitope-mapped using synthetic overlapping peptides in a bead-based array format for selection of clones with nonoverlapping epitopes only.
Isotyping
All monoclonal antibodies are isotyped to allow for multiplexing using isotype-specific secondary antibodies.
Hybridoma Cell Cultivation
Atlas Antibodies use in vitro methods for the production scale-up phase thus replacing the use of mice for production of ascites fluid.
Antibody Characterization
The characterization of Prestige Monoclonal Antibodies starts with an extensive literature search to select the most relevant and clinically significant tissues to use for IHC characterization. Often there are more than one tissue type displayed in the IHC application data for each antibody. In addition to positive stained tissue, a negative control tissue staining is also displayed and if relevant, clinical cancer tissue staining.
The Western blot (WB) characterization includes results from endogenous human cell or tissue protein lysates or optionally recombinant full-length human protein lysates.
Each monoclonal antibody is thus supplied with the most relevant characterization data for its specific target.
The product numbers of all Prestige Polyclonals start with ”HPA” and of monoclonal antibodies with “AMAB”.

Immunohistochemical staining of human breast tumour using Anti-HER2 (AMAb90627) shows strong membranous (combined with moderate cytoplasmic) positivity in tumour cells in HER2-positive ductal carcinoma, while HER2-negative ductal carcinoma shows no membranous positivity. By Western Blot analysis, HER2 is detected in the breast cancer cell line SK-BR-3.
Progesteron Receptor

IHC staining using the Anti-PGR antibody (HPA004751) in normal human corpus (uterine) tissue shows strong nuclear positivity in glandular cells. In the presented breast cancer sample, the staining of tumor cells is also nuclear. ICC-IF shows nuclear staining in U-251MG cells.
Estrogen Receptor

The Anti-ESR1 antibody (HPA000449) shows distinct nuclear positivity in glandular cells in human breast tissue and in tumor cells in breast cancer samples using IHC.

IHC staining using the Anti-ESR1 antibody (HPA000450) shows strong nuclear positivity in glandular and stromal cells of human corpus, uterine tissue and in tumor cells in breast cancer.
Ki67

The Anti-MKI67 antibody (HPA000451) shows strong nuclear positivity in a fraction of cells in the reaction center in human lymph node using IHC. In breast cancer, the staining of tumor cells is also nuclear and by ICC-IF, staining of the human cell line U-2OS shows positivity in nucleoli.

IHC staining of human tonsil tissue using the Anti-MKI67 antibody (HPA001164) shows nuclear staining of reaction center cells. In tumor cells in breast cancer, the staining is mainly nuclear and in U-2OS cells, using ICC-IF, nucleoli show strong positivity.
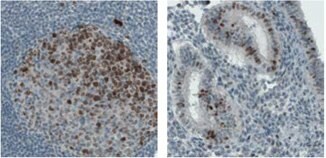
IHC staining of lymph node in human colon shows strong nuclear and nucleolar immunoreactivity in the reaction centrum cells using the monoclonal Anti-MKI67 antibody (AMAb90870). In uterus, nuclear positivity in a subset of glandular cells is shown.
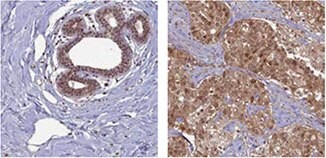
The Anti-BRCA1 antibody (HPA034966) shows positivity in glandular cells in normal human breast tissue and in tumor cells in breast cancer samples using IHC.

IHC staining using the Anti-BRCA2 antibody (HPA026815) in normal human breast tissue shows positivity in glandular cells. In breast cancer, nuclear staining of tumor cells is shown.
ACAT1

Immunohistochemical staining of human liver tissue using Anti-ACAT1 (HPA004428) shows strong cytoplasmic positivity in hepatocytes. By Western blot analysis, ACAT1 is detected in the human cell lines RT-4 and U251-MG and in liver and tonsil tissue lysates. By ICC-IF in the human cell line A-431, positivity is shown in mitochondria.
CD44

Immunohistochemical staining of human esophagus tissue using Anti-CD44 (HPA005785) shows strong strong cytoplasmic and membranous positivity in squamous epithelial cells. By Western Blot analysis, CD44 is detected in the human cell line U-251MG. ICC-IF in the human cell line U-251MG shows positivity in plasma membrane.
* WB both in human and rodent samples
References
Antibodies Against Gene Products in MammaPrint, Oncotype, EndoPredict and uPA Tests
This section presents antibodies in Prestige Antibody product catalog against gene products included in the diagnostic MammaPrint, EndoPredict, Oncotype and uPA tests. MammaPrint is a gene expression profile test based on the Amsterdam 70-gene breast cancer gene signature marketed by Agendia. It is a test to assess the risk that a breast tumor will metastasize to other parts of the body. MammaPrint aims at stratifying patients into “Low Risk” and “High Risk”. Oncotype DX (developed by Genomic Health) is the most frequently used gene expression profile in clinical practice in the United States analyzing a panel of 21 genes within a tumor to determine a Recurrence Score.
BIRC5/Survivin

The Anti-BIRC5 antibody (HPA002830) shows nuclear positivity in germinal center cells in human tonsil tissue and in tumor cells in colorectal cancer using IHC.
CD68/Macrosialin

IHC staining of human lung tissue using the Anti-CD68 antibody (HPA048982) shows strong cytoplasmic positivity in macrophages and in hematopoietic tissues, such as spleen.
DTL

IHC staining of human bone marrow using the Anti-DTL antibody (HPA028016) shows strong nuclear positivity in bone marrow poietic cells. By ICC-IF, staining of nucleus in U-251 MG cells is detected.
GSTM5

The Anti-GSTM5 antibody (HPA048652) shows cytoplasmic positivity in glandular cells in human rectum by IHC and in WB, the antibody detects a band of predicted size in cell lysates of RT-4, U-251 MG, as well as in liver tissue lysate.
* WB both in human and rodent samples
MMP9
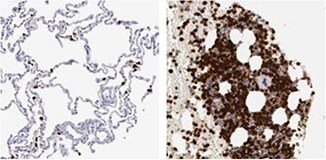
IHC staining of human lung tissue using the Anti-MMP9 antibody (HPA001238) shows strong nuclear positivity in macrophages and in bone marrow poietic cells in bone marrow tissue.

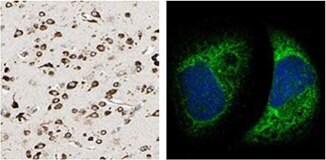
IHC staining using the Anti-MTDH antibody (HPA010932) shows strong cytoplasmic positivity in neuronal cells in human cerebral cortex tissue. In ICC-IF in A-431 cell line, the antibody stains endoplasmic reticulum.

IHC staining using the monclonal Anti-MTDH antibody (AMAb90762) shows strong cytoplasmic reactivity in tumor cells from breast and colorectal cancer samples.
P5C Dehydrogenase/ALDH4A1

IHC staining using the Anti- ALDH4A1 antibody (HPA006401) shows strong cytoplasmic positivity with granular pattern in human kidney and liver tissues.
* WB both in human and rodent samples
PITRM1/MP1

The Anti- PITRM1 antibody (HPA006753) shows strong cytoplasmic positivity in myocytes in human heart muscle using IHC. ICC-IF staining of human cell line U-251 MG shows positivity in mitochondria.
PRC1

IHC staining of human testis tissue using the Anti-PRC1 antibody (HPA034521) shows strong nuclear positivity in cells of seminiferus ducts. ICC-IF shows staining of nucleus, plasma membrane and microtubules in A-431 cells.
SCOT/OXCT1

IHC staining of human heart muscle and kidney by Anti-OXCT1 antibody (HPA012047) shows strong cytoplasmic positivity in myocytes and cells in tubules, respectively. ICC-IF shows staining of mitochondria in A431 cells.
* WB both in human and rodent samples
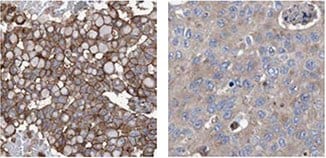
IHC analysis using Anti-KLHL26 antibody (HPA023074) shows a varying membranous/cytoplasmic staining pattern in breast tumor samples from different patients.

The Anti-ACSF2 (HPA024693) antibody shows granular cytoplasmic positivity in breast tumor cells from different patients varying from strong to negative.
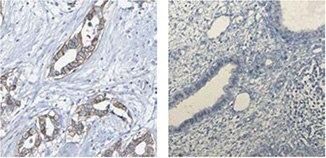
The Anti-GCM1 (HPA011343) antibody shows membranous positivity in breast tumor cells while normal breast tissue is negative.
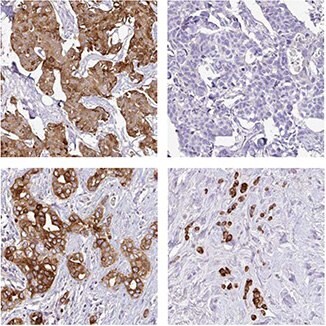
The Anti-AGR3 (HPA053942) antibody shows strong cytoplasmic positivity in 11/12 breast cancer patients, while 1 patient is completely negative.
* WB both in human and rodent samples
Finding Cancer Biomarkers
Breast Cancer
Breast cancer is the second most common cancer and by far the most frequent cancer among women. The incidence of breast cancer is increasing steadily, but without a corresponding increase in mortality. If detected at an early stage, the prognosis is relatively good for a patient living in a developed country, with a general five-year survival rate of approximately 85%.
Breast Cancer and Treatment
Cancer, though often denoted as a singular disease, is truly a multitude of diseases. This understanding has evolved over the years, but many patients are not receiving optimal treatment for their disease. For cancer patients to receive a more individualized treatment, there is still a need for new and better ways to stratify patients. The classical prognostic factors such as stage and grade of the tumor are insufficient for a correct estimation of patient prognosis. Additional information from cancer biomarkers promise to substantially improve this estimation, ultimately leading to a more individualized treatment, thus avoiding both under and overtreatment of patients.
The primary curative treatment for breast cancer patients is surgery, often in combination with adjuvant therapy. However, adjuvant therapy is associated with substantial costs and sometimes severe side effects, and physicians have identified reduction of overtreatment as the major clinical need in breast cancer treatment today. Thus, the stratification of patients into different prognostic categories is of great importance as it may aid physicians in selecting the most appropriate treatment for a given patient.
The majority of breast cancers are hormone receptor responsive, i.e., express the estrogen receptor (ER) and/or the progesteron receptor (PR). Patients with tumors expressing these receptors may receive adjuvant endocrine treatment, such as tamoxifen.
Breast cancers may also express the HER2 protein (human epidermal growth factor receptor 2), and patients with tumors expressing this protein may receive adjuvant therapy with trastuzumab.
Adjuvant treatment may also consist of chemotherapy or radiation therapy.
RBM3
The RNA-binding motif protein 3 (RBM3) is an RNA- and DNAbinding protein, whose function has not been fully elucidated. It has been shown that the protein is expressed as an early event in mild hypothermia, and also in other conditions relating to cellular stress, such as glucose deprivation and hypoxia1. During stress, RBM3 is thought to protect the cells by aiding in maintenance of protein synthesis needed for survival1. Recently, it has also been shown that RBM3 attenuates stem cell-like properties in prostate cancer cells2.
RBM3 was identified via the Human Protein Atlas (HPA) as a potential oncology biomarker through the differential expression pattern present in several cancers investigated as part of the HPA project (proteinatlas.org)3,4.
The IHC analysis using the Anti-RBM3 antibody HPA003624 showed a weak expression pattern in normal breast tissue, but a stratified pattern in breast cancer tissue (Figure 1). Researchers further investigated the expression in larger breast cancer cohorts and the expression of RBM3 was shown to be associated with a prolonged survival5.

Figure 1. Immunohistochemical analysis using the Anti-RBM3 antibody (HPA003624) shows weak expression in normal breast tissue (A) and differential expression, varying from weak to strong in tumor breast samples (B, C).
RBM3 as a Prognostic Biomarker in Breast Cancer
After identification of RBM3 as a potential prognostic biomarker, researchers further investigated the RBM3 protein expression in larger breast cancer cohorts1. In a cohort of 500 premenopausal women with stage II invasive breast cancer, RBM3 expression was found to be associated with small, low-grade, estrogen receptor (ER)-positive tumors. When analyzing the subset of ER-positive patients, RBM3 was an independent predictor of recurrence free survival (RFS). As shown in Figure 2, patients with tumors expressing high levels of the RBM3 protein have an improved survival compared to patients with tumors expressing low levels of RBM3.
RBM3 protein expression has further been analyzed in many different patient cohorts from various forms of cancer. Levels of RBM3 expression was found to have a significant connection to patient survival in breast1, colon2, ovarian3,4, testicular, urothelial5, and prostate6 cancer as well as in malignant melanoma7.
In conclusion, RBM3 is a marker of good prognosis in breast cancer as well as in several other cancers.

Figure 2. Kaplan-Meier (survival) analysis of recurrence free survival (RFS) according to RBM3 expression for ER-positive breast cancer patients. Patients were split into two groups based on high and low RBM3 expression.
RBM3 Antibodies
There are two Anti-RBM3 antibodies in Atlas Antibodies' product catalog; the polyclonal HPA003624 and the PrecisA Monoclonal AMAb90655. The monoclonal Anti-RBM3 antibody AMAb90655 has shown excellent specificity in Western blot analysis of human cell lines, and is routinely used for staining of formalin fixed paraffin embedded tissue in IHC (Figure 3).

Figure 3. Immunohistochemical analysis of RBM3 expression in breast cancer (left) and prostate cancer (right) using AMAb90655 shows nuclear positivity in tumor cells. The WB image shows an expected band of 17 kDa in human cell line RT4 lysate using AMAb90655.
References
Granulin
Granulins are a family of secreted, glycosylated peptides that are cleaved from a single precursor protein. Cleavage of the signal peptide produces mature granulin which can be further cleaved into a variety of active peptides. These cleavage products are named granulin A, granulin B, granulin C, etc. Both the peptides and intact granulin protein regulate cell growth. Different members of the granulin protein family may act as inhibitors, stimulators, or have dual actions on cell growth. Granulin family members are important in normal development, wound healing, and tumorigenesis [provided by RefSeq, Jul 2008].
In a paper by Elkabets et al, the role of GRN expression in responding tumor instigation was investigated by studying recrution of GRNexpressing bone marrow cells into responding tumors in mice1. Certain tumors can foster the growth of other tumors or metastatic cells located at distant anatomical sites, which is referred to as tumor instigation. In this study, rigorously growing human breast carcinoma cells were implanted in mice and it was shown that these cells stimulated the outgrowth of otherwise poorly tumorigenic, indolent transformed cells. GRN was identified as the most upregulated gene in the instigating bone marrow cells. The GRN expressing cells induced resident fibroblasts to express genes that promoted malignant tumor progression. It was speculated whether anticancer therapies might involve targeting GRN, or the activated GRN expressing cells, and thereby disrupting these cell lines of communication that promote cancer progression.
By using the Anti-GRN antibody HPA028747 in the analysis of tumor tissues from a cohort of breast cancer patients, high GRN expression was shown to correlate with the most aggressive triple-negative, basal-like tumor subtype and reduced patient survival (Figure 4).
References
![GRN expression was shown to correlate with aggressive tumor subtypes and reduced survival of breast cancer patients using antibody HPA028747. The diagram to the left shows percentage of tumors in each category (Triple-Negative [TN]/ basal or nonbasal) that show high GRN positivity and the Kaplan-Meier analysis to the right shows correlation between GRN-positive (green) or GRN-negative (blue) expression and survival. grn-expression](/deepweb/assets/sigmaaldrich/marketing/global/images/technical-documents/articles/research-and-disease-areas/cancer-research/grn-expression/grn-expression.jpg)
Figure 4. GRN expression was shown to correlate with aggressive tumor subtypes and reduced survival of breast cancer patients using antibody HPA028747. The diagram to the left shows percentage of tumors in each category (Triple-Negative [TN]/ basal or nonbasal) that show high GRN positivity and the Kaplan-Meier analysis to the right shows correlation between GRN-positive (green) or GRN-negative (blue) expression and survival.

IHC staining of human pancreas tissue using the Anti-GRN antibody (HPA008763) shows strong cytoplasmic positivity in exocrine glandular cells. ICC-IF shows positivity in vesicles in A-431 cells.

IHC analysis using the Anti-GRN antibody HPA028747 shows strong cytoplasmic positivity in normal duodenum tissue in glanduclar cells and vesicle positivity in U-251 MG cells.
Anillin
Anillin is an actin binding protein that is a subunit of microfilaments, one of the cytoskeleton components. Anillin is expressed in most cells and is involved in basic cell functions, e.g. motility, division and signaling. Studies of anillin expression have shown that it is over expressed in several human tumors.
Anillin as a Treatment Predictive Prognostic Biomarker in Breast Cancer
Anillin expression was analyzed in a patient cohort consisting of 467 samples from patients diagnosed with breast cancer, using the Anti-ANLN antibody HPA005680. Patients with tumors expressing high levels of anillin had a reduced recurrence free survival (RFS) compared to patients with tumors expressing low levels of anillin (Figure 5A). The same association between anillin expression and reduced survival could be seen when analyzing breast cancer specific survival (BCSS, Figure 5B). In a study by O´Leary et al, the prognostic impact of anillin was confirmed by Cox regression analysis. High anillin expression was associated with reduced BCSS and RFS in univariate- as well as in multivariate analysis, adjusted for tumor size and grade, age at diagnosis, nodal-, ER-, PR-, HER2-, and Ki67 status.
In conclusion, anillin is a marker for poor prognosis in breast cancer.

Figure 5. Kaplan-Meier (survival) analysis of recurrence free- (A) and breast cancer specific survival (B) according to aniliin expression for breast cancer patients. Patients were split into two groups based on high and low anillin expression.
References
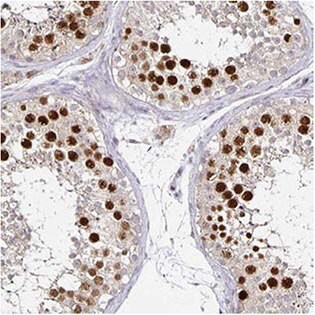

The Anti-ANLN antibody (HPA005680) shows strong nuclear positivity in cells in seminiferous ducts in human testis by IHC. In ICC-IF, nuclei (but not nucleoli) of A-431 cells stain positively and in WB, the antibody detects a band of predicted size in cell lysates of RT-4 and U-251 MG.

Anti-ANLN antibody AMAb90660 shows strong nuclear immunoreactivity in a subset of tumour cells in lung adenocarcinoma and a band of predicted size in human cell line U-251 MG.

AMAb90662 Anti-ANLN antibody shows strong nuclear immunoreactivity in a subset of tumor cells in colorectal cancer and a band of predicted size in human U-251 MG cells.
Pour continuer à lire, veuillez vous connecter à votre compte ou en créer un.
Vous n'avez pas de compte ?