Impact of GenElute™-E Purification Method on Accuracy of DNA Quantitation and Downstream Enzymatic Processes
Such bind-wash-elute methods are tedious, requiring multiple wash and spin steps. Repeated spin steps can result in significant DNA shearing. Additionally, chaotropic salts and other contaminants can easily carry through into the eluted DNA or RNA, affecting final purity and quantitation as well as downstream enzymatic processes, such as PCR.
We evaluated a novel single-spin nucleic acid purification system that eliminates the need for high salt binding and ethanol wash steps. GenElute™-E DNA and RNA purification kits employ a negative chromatography method dependent on size exclusion to separate large DNA and RNA nucleic acid molecules from smaller protein, lipid, and ionic components in cell, tissue, blood, and other samples (Figures 1-2).

Figure 1. Purification of nucleic acid using negative chromatography (size exclusion) techniques.

Figure 2.Elution of purified DNA and retention of proteins, lipids, and other molecules using and GenElute™-E Single Spin Blood DNA High Yield Kit with mouse blood samples. A) Absorbance spectra of purified genomic DNA (red line). B) Absorbance spectra of GenElute™-E spin column retentate (black line) overlaid on purified DNA spectra (red). Retentate spectra shows protein and other contaminants with OD at ~280nm or ≤240nm. Note change of scale on y-axis.
We evaluated the impact of nucleic acid purification method on impurities interfering with quantitation and downstream enzymatic processes. For these comparison studies, genomic DNA was prepared using GenElute™-E single-spin purifications technology or classic silica-based bind-wash-elute spin prep technology from Supplier X. Nucleic acid purity was assessed using three methods:
- UV spectrophotometry (optical density ratio as measured by A260nm/280nm and A260nm/230nm)
- Gel electrophoresis
- Quantitative PCR (qPCR)
Results indicate that GenElute™-E single-spin DNA and RNA purification systems provide higher purity than current silica spin prep techniques, leading to more accurate quantitation and decreased interference from contaminants in downstream enzymatic processes such as PCR.
Evaluation of Impurities Introduced During Nucleic Acid Preparation
Interference with Quantitation by Ultraviolet (UV) Spectrophotometry
Common biological contaminants, such as proteins, fragmented nucleic acids, ssDNA, RNA (if measuring DNA), primers, and chaotropic salts from extraction and purification protocols can lead to overestimation of nucleic acid concentration. Some of these impurities can be measured using ultraviolet (UV) spectrophotometry. In this method, UV light is passed through the purified DNA or RNA sample. The absorbance of the sample at various wavelengths is measured.
Absorbance at 260 nm (A260) is used to measure nucleic acid. Absorbance at 280 nm (A280) is used to measure contaminating protein in the sample. The A260/A280 ratio can be used to estimate sample purity with respect to protein contaminants. High purity DNA will have an A260/A280 ratio ≥ 1.8. A lower ratio indicates protein contamination in the final prep.
Absorbance at 230 nm (A230) can be used to identify the presence of chemical contaminants such chaotrophic salts. The A260/A230 ratio should ideally be greater than 2.0 for minimal chemical interference in enzymatic processes such as PCR. It is critical to assess this ratio when high purity samples are required.
The absorbance spectrum, A260/280 ratio, and A260/230 ratio were evaluated for GenElute™-E single-spin DNA and RNA purification systems using blood samples. Results were compared to those obtained from silica spin prep purification of the same samples using a competing product (Figure 3 and Table 1). The data suggest that GenElute™-E purification systems provide higher genomic DNA purity with fewer biological and chemical contaminants in the final sample preparation compared to silica bind-wash-elute preparation methods.

Figure 3. UV absorption spectra of genomic DNA prepared from blood using A) silica-based DNA purification spin prep kit from Supplier X or B) GenElute™-E single-spin negative chromatography DNA purification kits. Inset graph measured at 10X concentration of eluted fraction. Impurities are detected solely in the silica elution fractions. Black lines show spectra for sample purified using the indicated method with any process contaminant impact. Red lines are spectra for the purified sample without process contaminants, used as baseline controls. Orange lines are spectra for the purification method without using an actual sample, showing process contaminant impact on the spectrophotometric reading.
Sheared DNA and Low Molecular Weight Nucleic Acid Impurities Evaluated by Gel Electrophoresis
Size separation by gel electrophoresis provides a visual and quantitative assessment of sample purity. Larger genomic DNA migrates more slowly than smaller DNA and RNA fragments. The resulting bands can be observed or quantitated using a fluorescent dye.
Purified DNA samples prepared from mouse liver tissue using silica-based spin prep kits (Supplier X) or negative chromatography by size exclusion (GenElute™-E single-spin DNA purification kits) were separated on an agarose gel. SYBR green was used as a nucleic acid stain. To normalize results, the same mass of DNA from each purified sample was loaded into each lane of the gel. Visualization of the agarose gel on a transilluminator showed that darker, single bands of genomic DNA were obtained using negative chromatography (size exclusion) compared to the silica-based purification method (Figure 4).
Samples eluted from silica membrane spin columns showed fainter bands at the lower end of the gel, consistent with contaminating small RNA or sheared DNA molecules in the final prep. Because gel loading was normalized against DNA in the sample (as determined by absorbance at 260 nm), these data suggest genomic DNA yield is overestimated by this method as contaminating smaller nucleic acid molecules also contribute to absorbance measurement. The higher intensity single band of DNA obtained using the GenElute™-E negative chromatography (size exclusion) method suggests greater purity of genomic DNA from these tissue samples. Results were quantified using fluorometric methods (Figure 5).
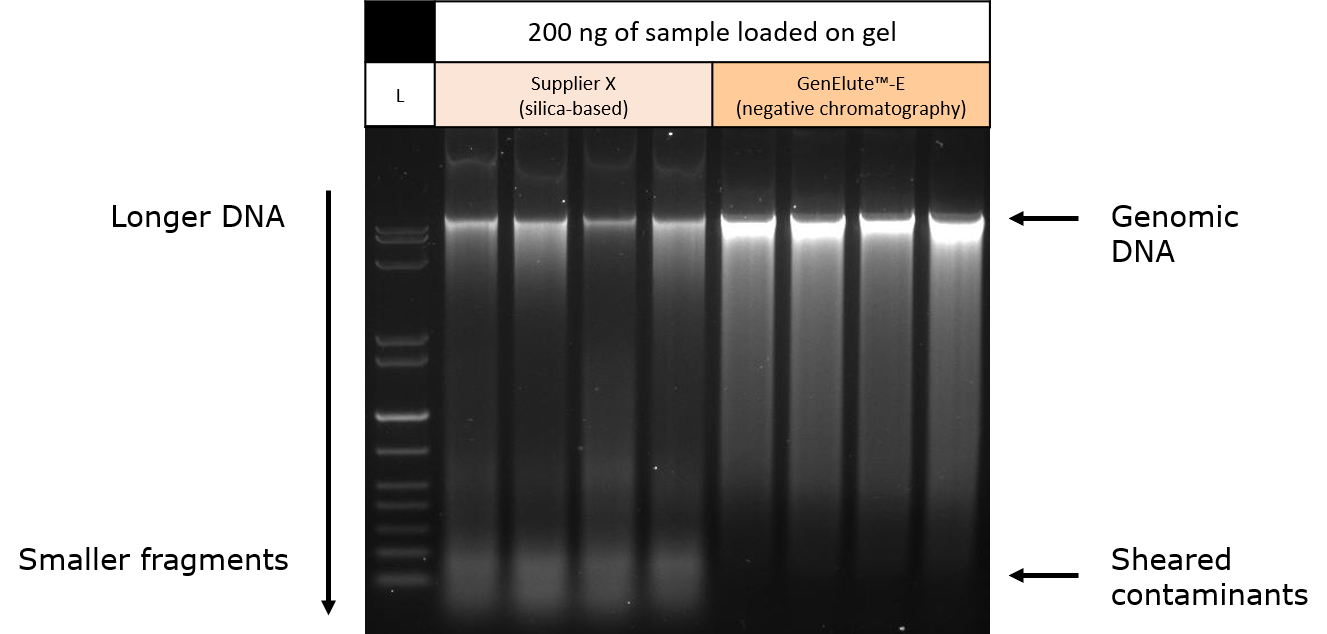
Figure 4. Gel electrophoresis of purified genomic DNA from mouse liver tissue samples. Mouse tissue was purified utilizing silica-based spin prep methods (Supplier X) or negative chromatography via size exclusion (GenElute™-E single-spin DNA purification kits). Samples were adjusted so that the same amount of DNA was loaded into each lane of the agarose gel. Results suggest the presence of contaminating RNA or sheared DNA in the samples purified using the silica-based method.

Figure 5. Calculated yield of genomic DNA (gDNA) and amounts of contaminating RNA or sheared DNA from mouse liver tissue samples. Samples were purified using silica-based spin prep methods (Supplier X) or negative chromatography via size exclusion (GenElute™-E single-spin DNA purification kits). Amounts of DNA were calculated by quantitating the fluorescent signal from the SYBR green stain. Results show small but significant amounts of RNA or sheared DNA contaminants in the samples prepared using silica-based purification spin columns.
Interference in qPCR
Silica-based purification methods introduce denaturing salts and organic solvents such as ethanol to the sample being purified. These contaminants can easily transfer to downstream applications and lead to inhibition of enzymatic reactions. Removal of such contaminants can render enzymatic methods such as qPCR more sensitive and robust.
To evaluate the presence of contaminants that inhibit enzymatic processes, genomic DNA from mouse kidney samples was purified using each spin purification method. Final samples were used in qPCR analysis experiments (Figure 6). The gene corresponding to beta-actin was also amplified as an endogenous control. Data show that the amplification curves for silica-purified samples were right-shifted compared to samples prepared using the GenElute™-E negative chromatography (size exclusion) purification method. This suggests the presence of interfering contaminants or lower than calculated yield in the silica-purified samples.

Figure 6. qPCR analysis for the determination of interfering contaminants in genomic DNA preparations. A) Amplification curves for genomic DNA prepared from mouse kidney tissue using a silica-based spin prep kit from Supplier X (blue curves) and GenElute™-E negative chromatography (size exclusion) spin prep kit (orange curves). Curves for the silica-purified samples are right-shifted compared to the GenElute™-E-purified samples, suggesting the presence of interfering contaminants or overestimation of starting concentration in the silica preps. B) Cq calculations using the β-actin gene as an endogenous control reference.
Conclusions
GenElute™-E single-spin DNA and RNA purification kits provide a convenient negative chromatography method for nucleic acid purification based on size exclusion. These kits offer an alternative to traditional silica-based spin preps, with improved purity as evidenced by UV spectrophotometry, gel electrophoresis, and qPCR.
Pour continuer à lire, veuillez vous connecter à votre compte ou en créer un.
Vous n'avez pas de compte ?