Downstream Applications for Plasmid DNA and Substrates
The purity of plasmid DNA in terms of protein, salt, particulates, RNA, and endotoxin contamination, and quality measures such as the percentage of supercoiled plasmid DNA, will all affect downstream functional performance. In general, plasmid DNA extracted using anion exchange-based systems is of higher quality and purity compared with that isolated using silica membrane-based systems. Table 5.5 provides a summary of downstream applications for plasmid DNA and substances that may interfere in each.
Restriction endonucleases
Enzymatic processing of purified plasmid DNA is one of the most common downstream applications. Restriction endonucleases are used to prepare DNA fragments for subcloning experiments (including the screening for the correct recombination event). Several restriction endonucleases are particularly sensitive to elevated salt levels (e.g., HindIII and SacI); digestion with these salt-sensitive enzymes can be used as a very broad indicator of contamination levels. Organic solvents affect enzyme activity, and RNA can interfere with visualizing digestion products on an agarose gel.
Plasmid DNA isolated with either QIAprep™ or illustra Mini Spin Kit was digested to completion in all the restriction digests performed in this study (Figure 5.2). In addition, digests involving low concentrations of the restriction enzyme HindIII (1 unit at 37 °C for 1 h) were performed. HindIII activity is diminished in the presence of elevated salt concentrations; therefore, HindIII digestion can be utilized to indicate the presence of prohibitively high salt contamination in the purified plasmid DNA. We observed that plasmid DNA samples isolated with both kits were digested to completion with HindIII (data not shown), suggesting that both illustra and QIAprep Kits produced plasmid DNA samples with negligible salt content.

Figure 5.2.Restriction enzyme digestion of plasmid DNA samples (400 ng, 5 units, 37°C for 1 h).
Modifying enzymes
Plasmid DNA can also be manipulated by a large and diverse range of modifying enzymes. In traditional ligation and cloning, the endonuclease digestion of plasmid DNA is commonly used to facilitate the subcloning of a DNA fragment from one plasmid into an alternative. Site-specific endonucleases are used to “cut” at enzyme recognition sites flanking the DNA sequence of interest. The digestion products are subjected to agarose gel electrophoresis, and the desired fragments are purified using kits such as illustra PCR and Gel Band Purification Kit from Cytiva. This process is applied to both the DNA fragment of interest and the alternative plasmid. The latter may also be subjected to treatment with alkaline phosphatase, derived from sources such as calf intestines. This enzyme removes terminal 3’-phosphate groups, preventing the religation of DNA molecules possessing compatible termini in the presence of DNA ligase. Thus, this modifying enzyme facilitates the ligation of the DNA fragment of interest into the new or alternative plasmid.
As with DNA endonucleases, many modifying enzymes, such as phosphatases and ligases, are also extremely sensitive to contamination, especially elevated salt and protein levels. Therefore, both the quality and purity of the extracted plasmid DNA are important. When choosing a kit to perform plasmid DNA extraction, take into consideration the quality of plasmid DNA produced and the required sensitivity of downstream applications. Commercially available kits such as those of the illustra range are designed to purify plasmid DNA that can be routinely used in the majority of molecular biology applications.
DNA sequencing
Options for sequencing of cloned DNA include standard enzymatic sequencing (e.g., using T7 DNA polymerase and ddNTP “chain terminators”) based on the Sanger method (8) and cycle sequencing. Cycle sequencing is similar to PCR in that it utilizes a thermostable DNA polymerase to generate new DNA. However, in cycle sequencing amplification is linear rather than exponential. Both options for sequencing are usually performed with fluorescent nucleotides so the DNA fragments can be analyzed automatically after gel electrophoresis. However, manual sequencing using radiolabeled nucleotides is still an option.
DNA sequencing can be used to indicate the quality of the extracted plasmid DNA. Quality is determined by automated DNA sequencing and Phred analyses. The Phred program is designed to assign nucleotide bases to DNA sequence traces. Phred reads DNA sequence chromatograms and analyzes the peaks to identify bases. It also assigns quality or Phred scores to each base. After identifying bases, Phred examines the peaks around each base to assign a quality score. Quality scores range from 4 to 60, with higher values corresponding to higher quality. These scores are linked to error probabilities. For example, a Phred score of 40 corresponds to a 1 in 10 000 chance that the base is called incorrectly, that is, 99.99% accuracy.
In general, good-quality plasmid DNA with high purity is required for manual sequencing. This is related to the amount of plasmid DNA required for the sequencing reaction. In comparison, cycle sequencing is more tolerant of contamination since contaminants are effectively diluted in the reaction mix.
Sequencing reactions (Figure 5.3) were performed on plasmid DNA isolated using two commercially available mini-scale purification kits (illustra and QIAprep™) according to the manufacturers’ instructions. DNA sequencing was performed on an ABI™ 3100 automated DNA sequencer using the BigDye™ Terminator v3.1 Cycle Sequencing Kit. Marginally higher Phred20 (99% accuracy) scores are generated when plasmid DNA is extracted using the midi-scale kits (data not shown). This is indicative of the higher quality plasmid DNA associated with the midiscale anion exchange-based kits.

Figure 5.3.Mean autosequence Phred20 quality measurements (above) for high-copy-number plasmid DNA samples prepared using QIAprep Spin Miniprep Kit (Qiagen) and illustra plasmidPrep Mini Spin Kits. The numbers 1, 2, and 3 refer to the culture from which the plasmid DNA was isolated. Processed culture volume (in ml) is shown in parentheses.
PCR amplification
Purified plasmid DNA is commonly used as a template in PCR for the amplification of specific cloned DNA sequences, for example, to modify the 5’ and 3’ ends of a cloned gene of interest with new restriction sites to facilitate subsequent subcloning experiments. PCR is also used to screen for the correct recombination event in subcloning experiments. However, in this context transformed E. coli cells are generally used as the template, indicating the relatively robust nature of PCR (and Taq DNA polymerase) in the presence of contaminants for routine amplifications.
A wide variety of DNA polymerases derived from thermophilic organisms are commercially available, including rTaq (Cytiva), PfuTurbo™ Hotstart DNA Polymerase and Vent™ DNA Polymerase. All add dideoxy nucleotides to the 3’-OH end of a primer in a template-directed reaction. Pfu and Vent DNA polymerases possess a 3’ to 5’ exonuclease proofreading activity, which reduces the number of misincorporated nucleotides in an extending primer. The error rates for the nonproofreading Taq DNA polymerase and Vent DNA polymerase are 285 × 10-6 and 57 × 10-6, respectively (New England Biolabs).
Due to the amplification associated with PCR, small amounts of plasmid DNA can be used as a template. Therefore, contaminants are essentially diluted away. Even so, routine PCR is a relatively robust process and intact bacterial cells can be successfully used directly in a PCR reaction with little impact on amplification efficiency.
Plasmid DNA purified using illustra plasmidPrep Mini Spin Kit was used as the template in a PCR reaction to amplify a 1.2 kb product with the thermostable DNA polymerases described above. The results are shown in Figure 5.4. All polymerases successfully amplified the product. Marginally higher efficiency was observed using the nonproofreading enzyme rTaq.
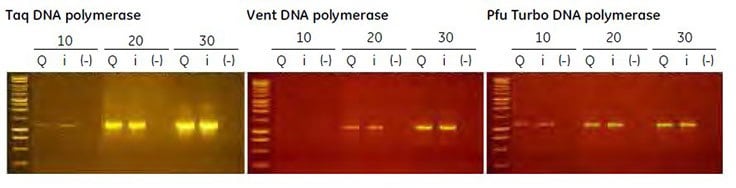
Figure 5.4.End-point PCR using several thermostable polymerases. Lanes Q and i represent amplification products derived from plasmid DNA samples extracted using QIAprep Spin Miniprep Kit (Qiagen) or illustra plasmidPrep Mini Spin Kit, respectively. (-) represents no template control reactions. The numbers 10, 20, and 30 indicate the number of thermal cycles performed. Aliquots (5 μl) of each reaction were loaded on a 1% (w/v) agarose gel.
Rolling circle amplification
In nature, the replication of circular DNA molecules, such as plasmids, occurs via a rolling circle mechanism. In the laboratory, linear rolling circle amplification (RCA) is the extension of an oligonucleotide primer annealed to a circular template DNA. Random hexamer primers are used to perform RCA of both strands of the plasmid, resulting in a cascade of strand displacement reactions producing an exponential amplification.
The application of RCA was demonstrated on plasmid DNA extracted using illustra plasmidPrep Mini Spin Kit. The RCA-amplified DNA generated was of sufficient purity and quality to successfully transfect a mammalian cell line, allowing the performance of a cell-based assay (data not shown).
Transfection
Purified plasmid DNA is routinely used for the transfection of mammalian or other eukaryotic cells. This process is very susceptible to contaminants (e.g., salt and endotoxin levels); therefore, the plasmid DNA must be of high purity and quality to ensure high transfection efficiencies and low toxicity. High-quality plasmid DNA is routinely isolated using many of the commercially available midi-/maxi-scale kits that are based on anion exchange, including illustra plasmidPrep Midi Flow Kit. The high-purity and -quality plasmid DNA associated with midi-scale kits is especially important for transfecting more sensitive cells such as primary cell lines and those that are highly sensitive to endotoxin levels. Certain robust cell lines (e.g., HEK293) can be transfected to relatively high efficiencies using plasmid DNA that is purified using silica-based mini-scale systems, including illustra plasmidPrep Mini Spin Kit. Mini-scale silica-based kits generally facilitate the purification of plasmid DNA that is of marginally lower quality in terms of salt and endotoxin levels compared with that extracted using anion exchange-based systems.
Plasmid DNA isolated using Qiagen™ Plasmid Midi and illustra plasmidPrep Midi Flow Kits was used to transfect the cell lines COS7, HeLa, and SH-SY5Y. The first two cell lines are representative of those that are relatively tolerant of contamination levels, while the latter SH-SY5Y cells are sensitive to elevated endotoxin levels. The results of these experiments are shown in Figure 5.5.

Figure 5.5.Transfection efficiencies of plasmid DNA generated using illustra PlasmidPrep Midi Flow Kit and Qiagen Plasmid Midi Kit. The numbers 25 and 50 indicate the culture volume (mL) from which plasmid DNA was extracted.
For COS7 and HeLa cell lines, both of the anion exchange-based midi-scale kits generated plasmid DNA of sufficient quality and purity to transfect the cells at comparable efficiencies (i.e., >70%). The endotoxin-sensitive SH-SY5Y cell line was transfected at comparable but lower efficiencies (i.e., ~10%), with plasmid DNA generated from both manufacturers’ kits.
Plasmid DNA isolated with illustra plasmidPrep Mini Spin and Midi Flow Kits was used to transfect the robust HEK293 cells. These cells are more amenable to transfection compared with the more sensitive cells described above. The highest transfection efficiency (~70%) was observed from plasmid DNA samples purified with illustra plasmidPrep Midi Flow Kit (50% efficiency was observed when using samples obtained from illustra plasmidPrep Mini Spin Kit). illustra Midi Flow Kit generates samples of high purity and quality that possess not only significantly reduced endotoxin levels, but also lower impurities such as proteins/salts/particulates compared with samples from illustra Mini Spin Kit.
To summarize, plasmid DNA purified using silica-based systems can be used to transfect certain robust cell lines. The advantage of using mini-scale extractions is that it facilitates the isolation and transfection of multiple samples relatively quickly and easily. Multiple plasmid DNA extractions based on a 96-well plate format that consists of silica membranes are commercially available and are routinely performed.
To continue reading please sign in or create an account.
Don't Have An Account?