Accelerating IAP-Based Protein Degrader Discovery with Novel Ligand Library

Targeted Protein Degraders
Protein degraders (e.g. PROTAC® degraders, SNIPERs) are emerging tools for therapeutic intervention by eliminating disease-causing proteins. A large class of protein degraders are bivalent small molecules that contain a ligand for an E3 ubiquitin ligase and a ligand for a target protein connected by a chemical linker (Figure 1). Thus, when added to cells, the degrader simultaneously binds the target and E3 ligase, triggering the polyubiquitination of the target protein and its subsequent degradation by the ubiquitin-proteasome pathway. Related are molecular glue degraders, which are smaller and more drug-like molecules that induce the interaction between an E3 ligase and target protein, which can be degraded by the same ubiquitin-proteasome degradation process. The direct degradation of target proteins provides multiple advantages over small-molecule inhibitors: for example, 1.) as degraders require only a binder (rather than an inhibitor that relies on deep enzymatic pockets), degraders may address >80% of the proteome considered “undruggable”); 2.) only sub-stoichiometric amounts are needed for potent activity; 3.) degraders are effective with low systemic exposure, which reduces off-target and toxic side effects.1
*PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.
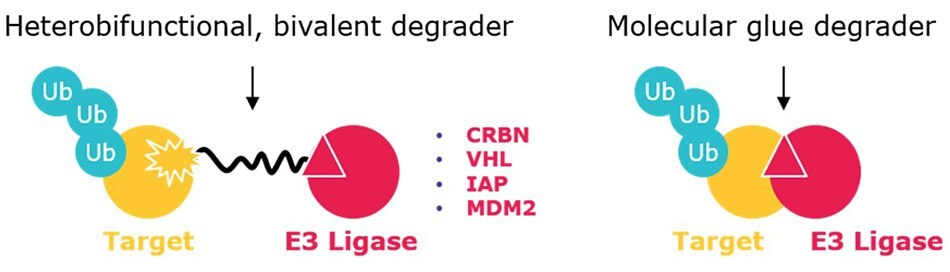
Figure 1.Heterobifunctional degraders (left) contain ligands for two different proteins – one typically being an E3 ubiquitin ligase – connected by a crosslinker. Molecular glues are smaller (non-conjugates) that include interactions between an E3 ligase and target (or neosubstrate).
Presently, fewer than 10 of the more than 600 E3 ubiquitin ligases have been exploited for targeted protein degradation. The most commonly utilized E3 ligases are VHL (Von‐Hippel‐Lindau ubiquitin ligase), CRBN (Cereblon), cIAP (cellular inhibitor of apoptosis protein1, and MDM2 (Mouse double minute 2 homolog). Utilitizing ComInnex expertise in library design and virtual screening, a novel IAP E3 ligand library was developed.
Generation of an AVPI-Mimetics IAP Library
The IAP family contains various proteins with high similarity (XIAP: X-linked IAP, cIAP1/2: cellular IAP1/2). XIAP inhibits initiator (Caspase-9) and effector caspases (Caspase-3 and -7) through the interaction of its BIR domains (BIR3 and BIR2) with the N-terminal tetrapeptide (AVPI) of the SMAC protein (Second Mitochondria-derived Activator of Caspases). Similarly, SMAC/AVPI can bind to the BIR3 domain of cIAPs and this interaction induces auto-degradation of the protein through ubiquitination.2 However, if SMAC mimetics (compounds having AVPI-like structural motifs) are linked to target-specific binders (“warheads”) in bifunctional degrader fashion, the conjugate may induce ternary complex formation of cIAP with the target protein, ubiquitination, and degradation. Auto-ubiquitination and targeted ubiquitination can be balanced by achieving binding selectivity towards XIAP vs. cIAPs.
AVPI (Ala-Val-Pro-Ile) is the minimum peptide sequence that is able to bind BIR3 domains of IAPs with various affinities (XIAP BIR3, cIAP1 BIR3 and cIAP2 BIR3 proteins with Ki = 3.6 μM, 184 nM, and 316 NnM, respectively).3 Based on the structure of this tetrapeptide, systematic research led to the discovery of potent, apoptosis-inducing IAP antagonists as well as E3 ligase-triggering SNIPERs (Specific and Non-genetic Inhibitor of apoptosis protein-dependent Protein ERasers) capable of degrading various targets (kinases, estrogen and retinoic acid receptors etc.).4
Virtual Screening of the AVPI-Mimetics IAP Library
ComInnex compiled a virtual E3 ligase ligand-like compound library containing around 100,000 AVPI mimetics that were in silico screened first. Then according to the SAR data a focused library of 168 compounds were designed and docked in the AVPI receptors (Figure 2). Good-resolution cIAP1 BIR3 structures were collected from the PDB database (4KMN, 3UW4, 4HY4, 4HY5, 4MTI, 4MU7, 4LGE). A cross-docking calculation was performed by docking all ligands into all receptors followed by MM-GBSA ΔGbind calculations. The 4LGE structure provided the highest average binding free energy; therefore, this structure was used for Induced-Fit Docking calculations. Upon virtual screening of the mimetics library, a smaller focused library of 148 members was obtained. The natural AVPI peptide (Figure 3a) showed -67.4 kcal/mol binding free energy, while replacement of Ile to Phe (AVPF, Figure 3a) resulted in a stronger binding (-71.2).
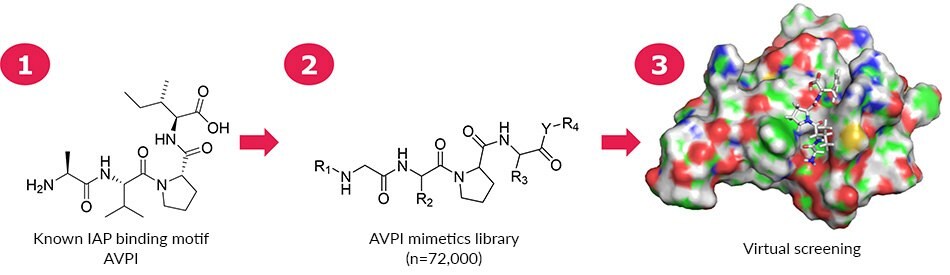
Figure 2.A 168-member mimetics focused library based on AVPI was assembled for virtual screening. On the right is shown the interaction between AVPF and 4LGE.
The 80 top-binding compounds (Figure 3) were compiled into an off-the-shelf library (Figure 4), which are selling as the ComInnex IAP Ligand Library, catalog # CIX001. In fact, AVPF-related architecture appeared in many potent SMAC mimetics and the lowest free energy (<-65 kcal/mol) members of the focused library are also mimicking this motif (Table 1). The Induced-Fit Docking and the subsequent MM-GBSA calculations were performed with the Schrödinger Small-molecule Drug Discovery suite (Schrödinger Release 2019-4; Glide, Schrödinger, llc, New York).
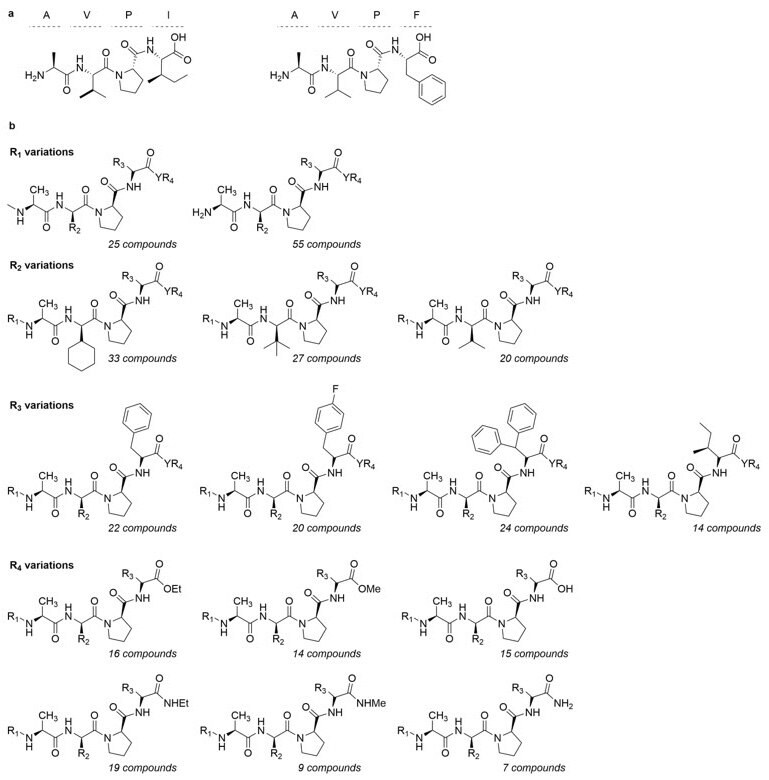
Figure 3.a) Structure of AVPI and AVPF. b) General structure of the AVPI-mimetics library. MM-GBSA calculations are shown in Table 1.
Applications of the IAP Ligand Library
Together, the in-silico leads represent novel structures in the IAP and targeted protein degradation fields, giving researchers an opportunity to develop chemically new IAP-mediated degraders with no IP hindrance. Each compound is provided at 125 µg solid material. Compound concentration in assays will vary depending on assay type, but common screening library stock solutions are 2 mM or 10 mM in DMSO. Please visit our ComInnex IAP Ligand Library Online Plate Map for structures and properties of all 80 compounds and details for ordering the individual compounds and synthetic derivatives for secondary assays or chemical optimization.
These novel compounds will find wide utility in IAP screening. For researchers interested in protein degradation, one may choose to screen for binding against IAP alone or in combination with a target. Based on results, leads may be developed into heterobifunctional degraders, molecular glue degraders, or chemical probes. Additionally, the leads may act as antagonists of the apoptosis inhibitor proteins to restore the inhibited apoptosis of the cancer cells. Here, an apoptosis screen would be performed with fluorescence indicators or readouts.
View of microrack |
Bottom view of microrack |
Individual vials |
Figure 4. ComInnex IAP Ligand Library
Accelerating IAP-Mediated Bifunctional Degrader Synthesis
The top three in silico leads (2075049, 2075085, 2075073 from the plate map) were derivatized into lead–linker conjugates for streamlined synthesis of heterobifunctional degraders akin to PROTACs and SNIPERs. As the modeling showed accommodation of the linker at both the N-terminus and C-terminus, a PEG or alkyl linker was added at each end to provide two different exit vectors and starting linker diversity. The amine terminal chemistry is compatible with our CRBN, VHL, and RNF4 ligand–linker conjugates to empower parallel synthesis from one starting target warhead in the generation of degrader libraries for screening. Read more in our technology spotlight here: Degrader Building Blocks with IAP In Silico-Derived Ligands.
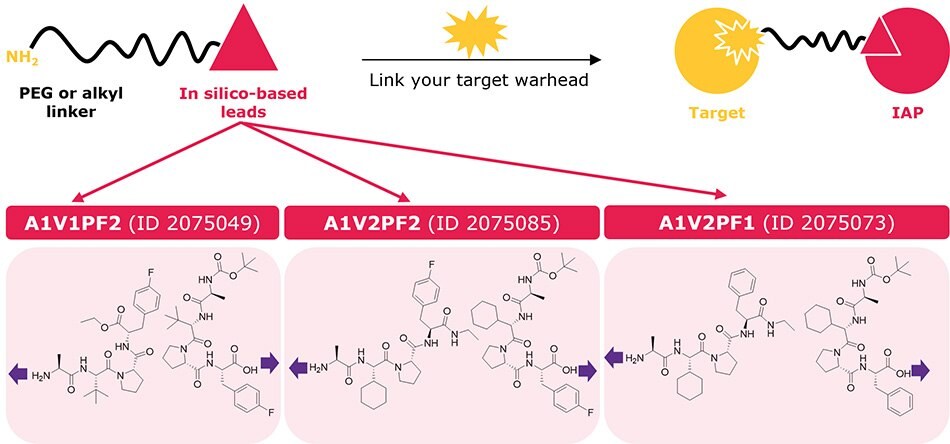
Figure 5.Ligand–linker conjugates containing the top three in silico-derived IAP leads (2075049, 2075085, 2075073) for synthesis of bifunctional degraders. Purple arrows indicate exit vectors where linkers are attached. Each lead ligand was conjugated at the N- or C-terminus with C6, C10, PEG1, and PEG3 linkers to offer 24 ligand–linker conjugates. Each lead ligand is also available individually (not conjugated to a linker) with a protected N- or C-terminus.
Table 1. Top 80 compounds and their MM-GBSA (kcal/mol).
Thank you to ComInnex for their partnership on this library and preparation of this technology spotlight.
References
To continue reading please sign in or create an account.
Don't Have An Account?