Genome Editing in Plants with CRISPR/Cas9
Successful ZFN-induced gene targeting was published as early as 2003. Since that time targeted genome editing technology has rapidly advanced and been made commercially available. Most recently, the discovery of the CRISPR/Cas9 pathway has accelerated interest in this field, opening up new possibilities for research and development. Although the CRISPR pathway was identified in bacteria as part of a putative adaptive immune system, it was quickly adapted to the purpose of modifying eukaryotic genomes. While tools like ZFNs laid the groundwork for genome editing today, there are limitations to this founding technology and others like it: the protein:DNA interaction of ZFNs makes designing them complex, the assembly of the ZFN expression construct is time-consuming, and the options for ZFN targeting are limited in many A-T rich plant genomes. The CRISPR pathway, as it has been adopted and modified for eukaryotic genome editing, overcomes many of these hurdles: it relies on an RNA:DNA interaction to find its genomic target, the recognition sequence required for this binding event is an easily altered 18-20 base pairs, and the only requirement for the nuclease binding is the presence of an NGG next to the target site. The first reports of the use of CRISPR/Cas9 in plants studying transient expression assays using Agrobacterium came out in 2013. The CRISPR/Cas9 technology has been successfully applied in model plants (Nicotiana benthamiana, Arabidopsis thaliana) and crops (rice, wheat) and the list is growing.
- CRISPR/Cas9: What is it and how does it work?
- Benefits of using CRISPR/Cas9 for genome-editing
- Simplified Workflow: CRISPR/Cas9 in Plants
- Ready-to-Use Cas9 and Guide RNA (gRNA) Expression Plasmids for Monocots and Dicots
- Custom CRISPR/Cas9 Plant Products
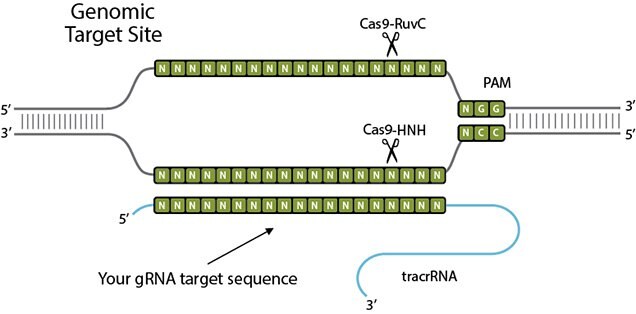
CRISPR/Cas 9: What is it and how does it work?
CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats. The discovery of the type II prokaryotic CRISPR “immune system” has allowed for the development for an RNA-guided genome editing tool that is simple, easy and quick to implement. The CRISPR/Cas9 system consists of a single monomeric protein and a chimeric RNA. A 20-nt sequence in the gRNA confers sequence specificity and cleavage is mediated by the Cas9 protein. Watson–Crick base pairing with the target DNA sequence is the basis for gRNA-based cleavage, making sophisticated protein engineering for each target unnecessary. Only a 20 nt in the gRNA is needs to be modified to facilitate the recognize a different target.
CRISPR/Cas9 consists of a Cas9 protein, a CRISPR RNA (crRNA), and a trans-activating crRNA ( tracrRNA). In gene editing applications, crRNA and tracrRNA are often fused into a single guide RNA (sgRNA). The ribonucleoprotein invades the target with crRNA guide sequence by forming a 20-bp RNA/DNA hybrid and displacing the opposite DNA strand after it encounters a protospacer adjacent motif (PAM), such as NGG. Cas9 endonuclease subsequently cleaves the complementary DNA strand (target strand) with a HNH nuclease domain and the displaced DNA strand (non-target strand) with a RuvC-like nuclease domain to create a double strand break (DSB). The repair of the DSB by host cell via non-homologous end joining (NHEJ) or homology directed repair (HDR) pathways can be utilized to create gene knockout or introduce a specific genetic modification through homologous recombination with a DNA donor.
RNA-guided endonucleases (RGENs) consisting of the Cas9 protein derived from Streptococcus pyogenes and guide RNAs (gRNAs) can be customized by replacing only the RNA component leading to decreases in labor and time compared to other gene editing methods. Using either Agrobacterium tumefaciens or by transfecting plasmids that encode them, programmable nucleases can be delivered into plant cells, where these nucleases cleave chromosomal target sites in a sequence-dependent manner. The result is site-specific DNA double-strand breaks (DSBs) whose repair by endogenous systems results in targeted genome modifications.
Benefits of using CRISPR/Cas9 for genome-editing
Main advantages of CRISPR/Cas9 are in terms of simplicity, accessibility, cost and versatility.
CRISPR/Cas9 system does not require any protein engineering steps, making it much more straightforward to test multiple gRNAs for each target gene.
Only 20 nt in the gRNA sequence need to be changed to confer a different target specificity, which means that cloning is also unnecessary.
Any number of gRNAs can be produced by in vitro transcription using two complementary annealed oligonucleotide. This allows the inexpensive assembly of large gRNA libraries so that the CRISPR/Cas9 system can be used for high-throughput functional genomics applications.
Another advantage of CRISPR/Cas9 compared to ZFNs and TALENs is the ease of multiplexing. The simultaneous introduction of DSBs at multiple sites can be used to edit several genes at the same time. It can be particularly useful to knock out redundant genes or parallel pathways. Researchers can engineer large genomic deletions or inversions by targeting two widely spaced cleavage sites on the same chromosome. Multiplex editing with the CRISPR/Cas9 system simply requires the monomeric Cas9 protein and any number of different sequence-specific gRNAs. In contrast, multiplex editing with ZFNs or TALENs requires separate dimeric proteins specific for each target site.
CRISPR/Cas9 system can cleave methylated DNA in human cells allowing genomic modifications that are beyond the reach of the other nucleases. While this has not been specifically explored in plants, it is reasonable to believe that the ability to cleave methylated DNA is intrinsic to the CRISPR/Cas9 system and not dependent on the target genome.
Simplified Workflow: CRISPR/Cas9 in Plants
Plant Biotechnology is entering a new era with the introduction of genome editing technologies that enables precise manipulation of specific genomic thereby superseding older methods of random mutagenesis as EMS mutagenesis and g-radiation sequences. Plant CRISPR/Cas9 products are intended for Agrobacterium-mediated plant transformation or biolistic microparticle bombardment or protoplast transformation. The products are based on the type IIA CRISPR/Cas9 derived from Streptococcus pyogenes. The native Cas9 coding sequence was codon optimized for expression in monocots and dicots, respectively. The monocot Cas9 constructs contain a monocot U6 promoter for sgRNA expression, and the dicot Cas9 constructs contain a dicot U6 promoter. The plant selection markers include hygromycin B resistance gene, neomycin phosphotransferase gene, and the bar gene (phosphinothricin acetyl transferase).

The pipeline of generating a CRISPR/Cas9-mutagenised plant line. c, control; m, mutagenized; RE, restriction enzyme. CELI and T7E1 are DNA endonucleases used in the surveyor assay.
Ready-to-Use Cas9 and Guide RNA (gRNA) Expression Plasmids for Monocots and Dicots
- Our plant CRISPR/Cas9 products are intended for Agrobacterium-mediated plant transformation or biolistic microparticle bombardment or protoplast transformation
- A codon optimized Cas9 protein and a gRNA are expressed from a single vector and provided as ready-to-use, transfection-grade DNA.

Basic structure of CRISPR/Cas9 constructs for Agrobacterium-mediated Transformation.

Basic structure of CRISPR/Cas9 constructs for biolistics or protoplast Transformation.
Plant CRISPR/Cas9 Product List |
|---|
Custom CRISPR GUS vectors |
|---|
References
Para seguir leyendo, inicie sesión o cree una cuenta.
¿No tiene una cuenta?